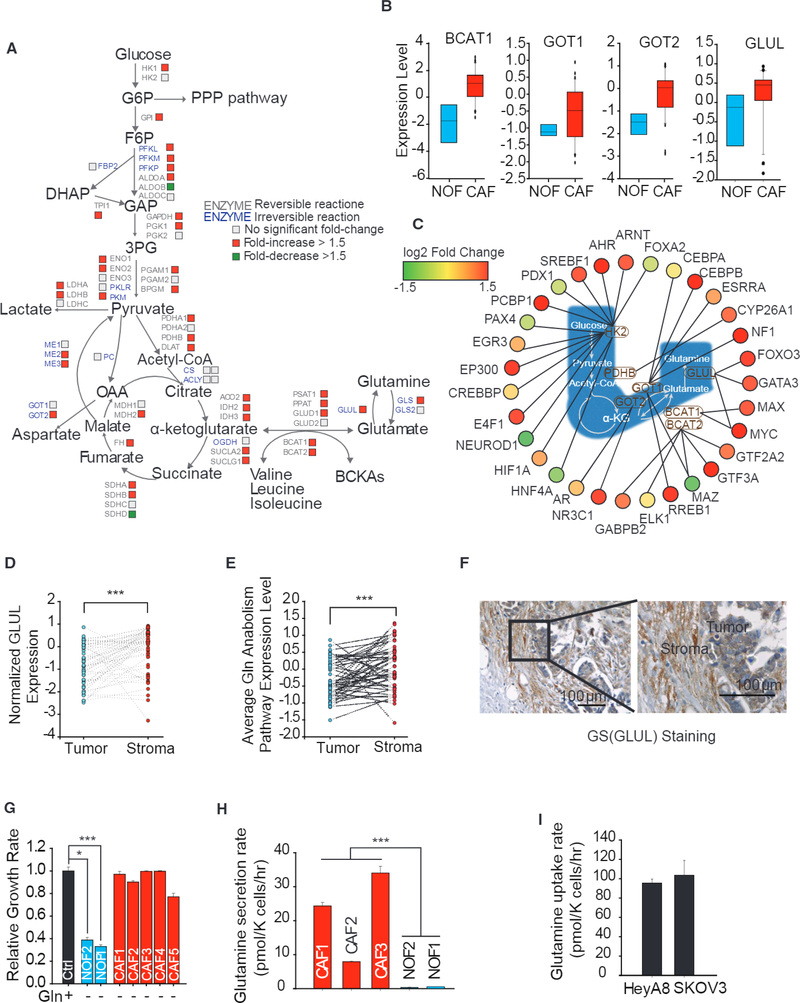
Figure 1. Gln Anabolic Pathway Is Upregulated in CAFs Compared to NOFs.
(A) Differential expression of genes encoding metabolic enzymes in cancer-associated fibroblasts (CAFs; n = 33) relative to normal ovarian fibroblasts (NOFs; n = 8). CAFs were derived from fibroblastic stromal components microdissected from a series of advanced-stage, high-grade serous ovarian adenocarcinomas; NOFs were derived from normal ovaries obtained from patients.
(B) Expression level of branch-chain aminotransferase 1 (BCAT1), glutamic-oxaloacetic transaminase 1 (GOT1), GOT2, and GS (GLUL) In CAFs and NOFs.
(C) Network of transcription factors regulating Gln anabolic and glycolysis pathway genes in CAFs relative to NOFs.
(D) GLUL expression levels in paired tumor epithelial and stromal compartments. Lines connecting tumor and stromal data points signify tumor and stromal samples derived from the same patient (Wisconsin test).
(E) Average pathway expression of Gln anabolism levels in paired tumor epithelial and stromal compartments (Wisconsin test).
(F) Representative IHC staining image comparing GS protein expression between stromal and tumor compartments.
(G) Proliferation after 72 hr of patient-derived NOF1, NOF2, and CAF1–5 under Gln deprivation relative to nutrient-rich media.
(H) Gln secretion rate of three patient-derived CAF1–3 and patient-derived NOF2/NOF1 under Gln deprivation conditions.
(I) Gln uptake rate of high-grade OVCA cell lines, HeyA8 and SKOV3, in Gln-replete medium.
Error bars indicate mean ± SEM of n ≥ 3 independent experiments.*p < 0.05, **p < 0.01, ***p < 0.001.