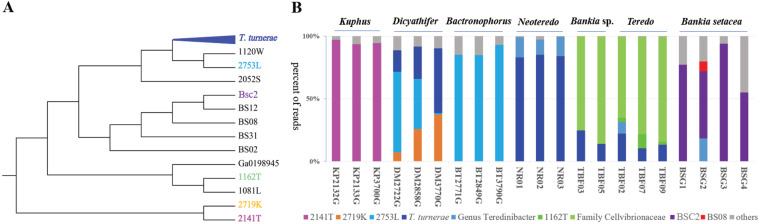
FIG 2.
Cultivated bacterial isolates represent the major shipworm gill symbionts. (A) Isolated bacteria analyzed in this study are shown in abstracted schematic of a 16S rRNA phylogenetic tree. The complete tree with accurate branch lengths and bootstrap numbers is shown in Fig. S1. T. turnerae comprised 11 sequenced strains; for other groups, individual strains are shown. Each color indicates different bacteria appearing in the metagenomes in panel B. (B) Species composition of shipworm gill symbiont community based on shotgun metagenome sequence analysis. The y-axis data indicate the percentages of reads mapping to each bacterial species, while the x-axis data indicate the individual shipworm specimens used in the study. Colors indicate the origin of bacterial reads; gray represents minor, sporadic, unidentified strains.