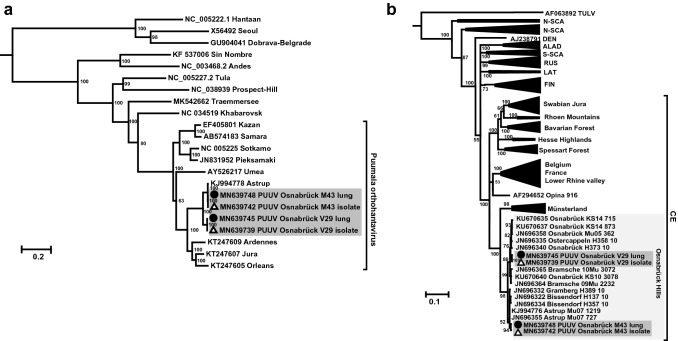
Fig. 4.
Hantavirus phylogenetic trees. Hantavirus phylogenetic tree of concatenated S, M, and L coding sequences of 18 published and four novel complete genomes (a). Partial S segment coding sequences of 365 nucleotides length, reconstructed with four novel and 202 published partial sequences (b). New PUUV isolates (GenBank accession numbers: MN639737-MN639742) are indicated with triangles and sequences derived from their original lung tissue (GenBank accession numbers: MN639743-MN639748) are labeled with black dots. Published sequences of other hantaviruses are labeled with GenBank accession numbers. Novel sequences are highlighted in gray. Posterior probabilities for major nodes of the maximum clade credibility phylogenetic tree are displayed. Analysis was performed by Bayesian algorithms via MrBayes v.3.2.6 (https://sourceforge.net/projects/mrbayes/files/mrbayes/) on the CIPRES online portal [28]. A mixed nucleotide substitution matrix was specified in 4 independent runs of 107 generations. Scale bar indicates nucleotide substitutions per site. For clarity, previously characterized PUUV clades from other parts of Europe are shown in simplified form. CE, Central European; LAT, Latvian; ALAD, Alpe-Adrian; S-SCA, South Scandinavian; N-SCA, North Scandinavian; RUS, Russian; FIN, Finnish; DEN, Danish