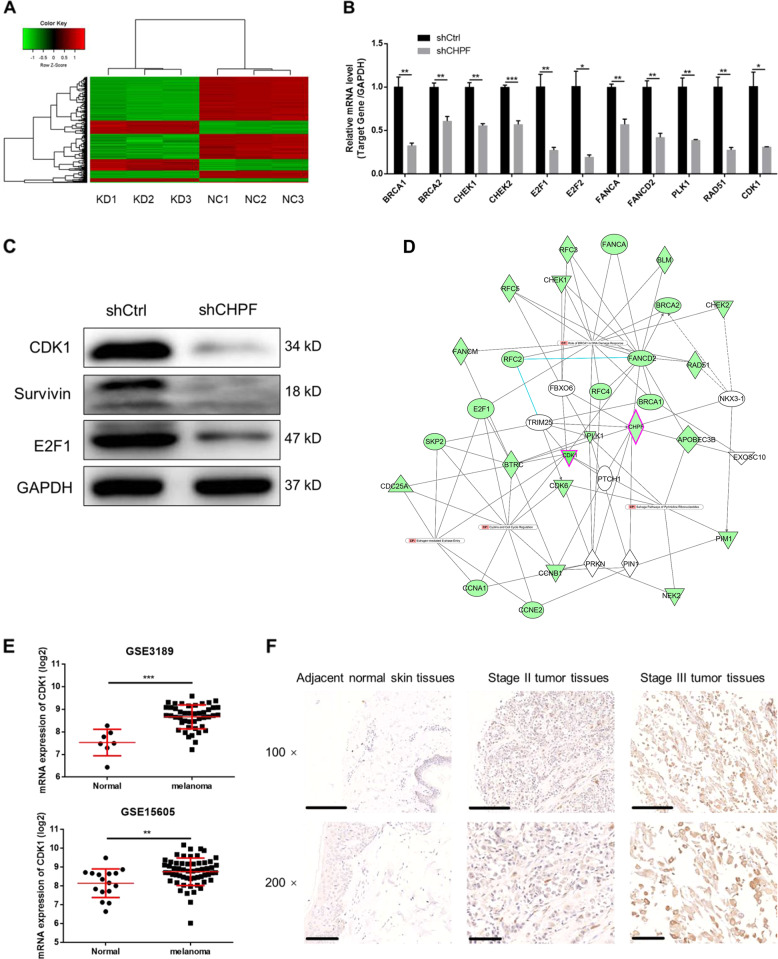
Fig. 4. Exploration of underlying mechanism by RNA-sequencing and IPA analysis.
a The heatmap of DEGs identified by RNA-sequencing of cells treated with shCtrl (n = 3) or shCHPF (n = 3). b IPA analysis of canonical signaling pathway was performed to identify the pathways regulated by CHPF knockdown. The dots were scaled by –Log (p value). The pathways in red were activated, and these in blue were inhibited. c The expression of several most significantly differentially expressed genes identified by RNA-sequencing, including BRCA1, CDK1, Survivin, and E2F1 was further detected in OM431 cells. d The IPA analysis of CHPF-related interaction network. e The upregulated expression of CDK1 in MM tissues was proved by data mining of GSE3189 and GSE15605 datasets. f The expression of CDK1 in normal skin tissue and stage II/III tumor tissues was detected by IHC analysis (Magnification 200×). *P < 0.05, **P < 0.01, ***P < 0.001.