Figure 1.
SPINK5 Disruption in Human Keratinocytes
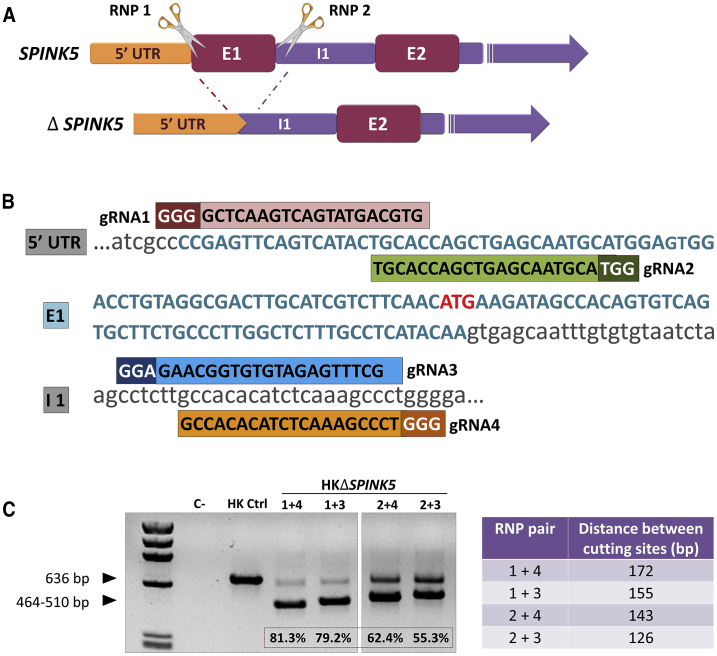
(A) SPINK5-knockout strategy. Pairs of CRISPR gRNA guides (RNP1 and RNP2) were designed to allow excision of SPINK5 exon 1 (E1) by Cas9. (B) Sequences and alignment of the designed gRNA guides to the targeted SPINK5 sequences (1 and 2 targeting E1, and 3 and 4 targeting intron 1 [I1]). The protospacer adjacent motif (PAM) is indicated in a darker color, and the transcription start site is indicated in red. (C) PCR analysis of genomic DNA from ΔSPINK5 keratinocytes treated with the different RNP pairs. Distance between cutting sites, according to each RNP pair combination, is shown in the right panel, and the efficacy (densitometric) value of excision is indicated at the bottom of each lane. The RNP 1+4 and 1+3 pairs yield the highest proportion of excised E1 according to the intensity of the lower band (464–510 bp).