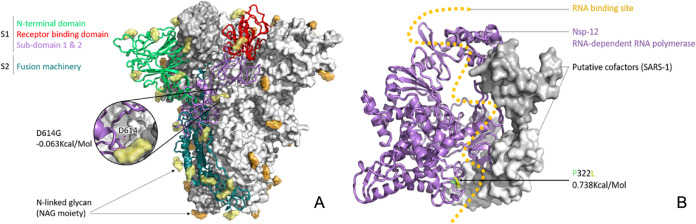
FIG 1.
(A) SARS-CoV-2 spike (S) trimer modelled with SWISS-MODEL (9) using 6VXX structure as a template, drawn and colored in PyMol (https://pymol.org/2/). Domains of a single S1 protomer are shown in cartoon view and colored green (N-terminal domain, NTD), red (C-terminal domain/receptor binding domain, CTD/RBD), and purple (subdomains 1 and 2, SD1 and SD2). S2 is shown in dark teal, while N-acetylglucosamine moieties are colored yellow (cartoon protomer) or orange (surface protomers). The enlarged inset shows the location of D614, which is where a mutation has arisen in the R03006/20 South African strain, buried in the interprotomer interface. (B) SARS-CoV-2 nsp12 (RNA-dependent RNA polymerase, RDRP) modelled with SWISS-MODEL (9), drawn and colored in PyMol, based on the nsp12, nsp7, and nsp8 protein complex of SARS-1 (PDB ID 6NUR). The RNA binding groove is indicated (orange), with the adjacent P322 (green) to L322 (yellow) mutation shown in stick view.