Figure 1.
miR-4500 Influences the Development of Breast Cancer through Regulation of the RRM2-Dependent MAPK Signaling Pathway
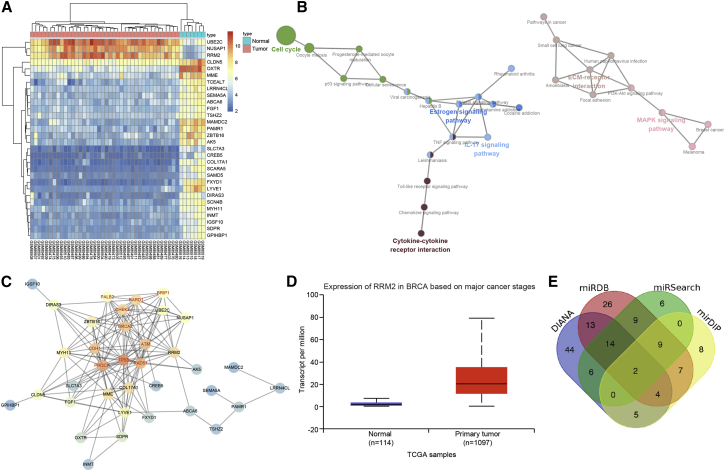
(A) The heatmap of variance analysis in breast cancer expression microarray data, wherein the abscissa represents the sample number, the ordinate represents the gene name, the upper dendrogram shows the sample-type clustering analysis, the ordinate represents the gene-expression level clustering, and each small square in the figure represents the expression level of a gene in a sample. (B) KEGG analysis of DEGs in breast cancer expression microarray data, wherein each circle represents a name of a disease or a name of KEGG entry, and the lines between the circles indicate their relationship. (C) The interaction network graph between known genes and DEGs in breast cancer, wherein each circle represents a gene, and the lines between the two circles indicate the interaction between the two genes. The red font represents the known genes related to breast cancer retrieved from the database. The circle color indicates the core degree of the gene in the network graph. The darker color reflects the higher core degree. (D) The expression analysis of the RRM2 gene in TCGA breast cancer database; the abscissa indicates the sample type, the ordinate indicates the gene-expression level, the left box shows the expression of RRM2 in normal samples, and the right box shows the expression of RRM2 in breast cancer samples. (E) The prediction results of regulatory miRNAs toward RRM2. The four ellipses in the figure represent the prediction results of four databases, and the middle part represents the intersection of them; besides, the numbers in the figure represent the number of miRNAs in each area.