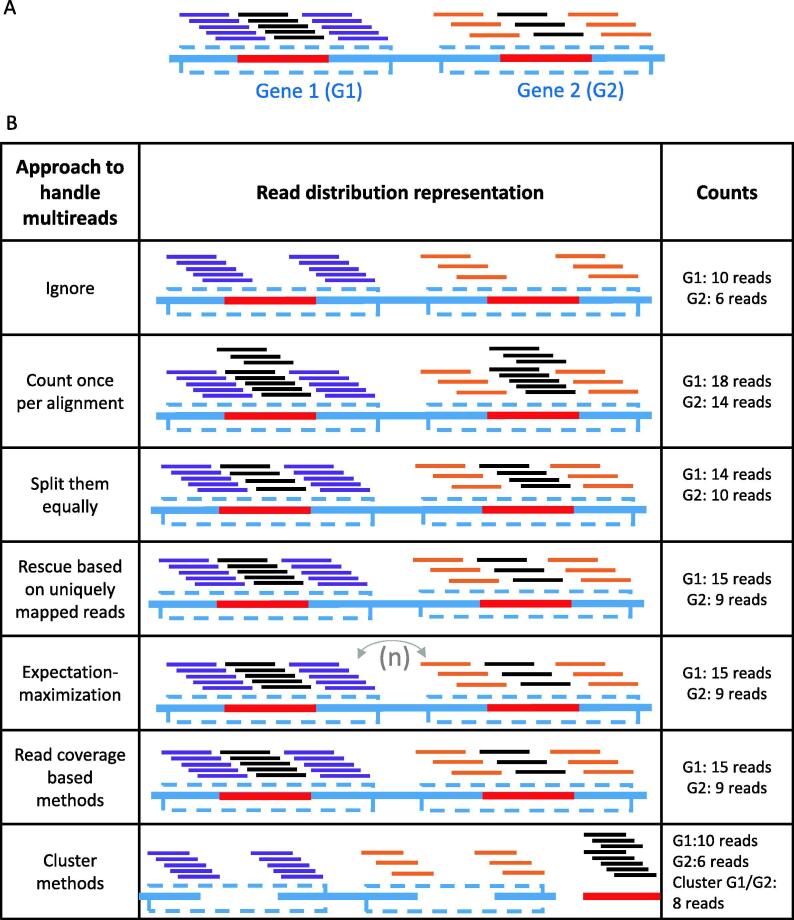
Fig. 3.
Strategies to deal with multi-mapped reads. (A) Example of two genes sharing a duplicated sequence and the distribution of RNA-seq reads originating from them. The two genes are represented by boxes outlined by dashed lines and their common sequence is illutrated by a red line. The reads are represented by lines above the genes, purple for reads that are unique to Gene 1, orange for reads that are unique to Gene 2 and black for reads that are common to genes 1 and 2. (B) General classes to handle multi-mapped reads include ignoring them, counting them once per alignment, splitting them equally between the alignments, rescuing the reads based on uniquely mapped reads of the gene, expectation–maximization approaches, rescuing methods based on read coverage in flanking regions and clustering methods that group together genes/transcripts with shared sequences. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)