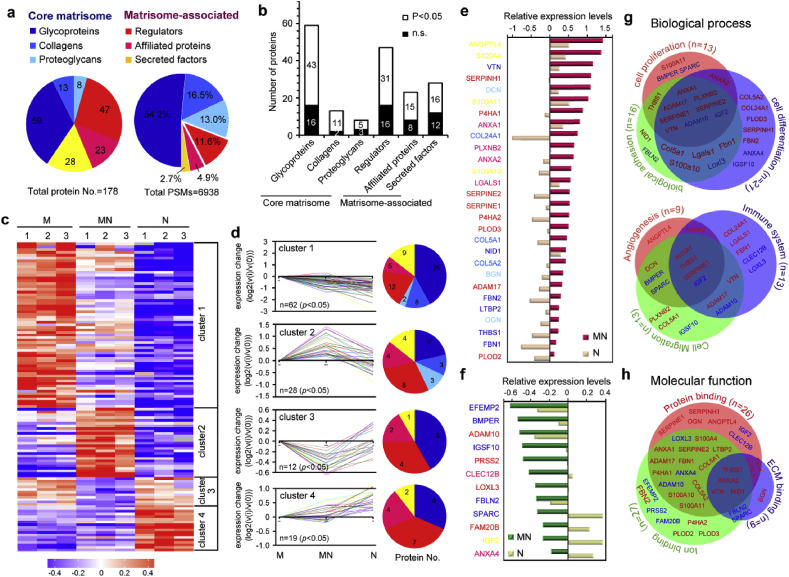
Fig. 6.
Quantitative proteomic analysis of ECMs secreted by cocultured cells (MN-ECM) revealed a unique matrisome signature. (a) Matrisome signature of cell secreted ECM as presented by pie chart. Distribution of ECM proteins was calculated by protein numbers (left) and protein abundance (PSMs, right) of each matrisome protein sub-category, respectively. (b) Distribution of protein numbers with or without statistical significance among three kinds of ECM. (c) Heatmap of quantitative proteomics using TMT labeling. The fold-change in protein detection levels normalized to the mean of M-ECM in log2 scale was shown. (d) Short-time series expression miner (STEM) of ECM proteins with statistical significance. According to the expression tendency, ECM proteins were divided into four clusters. Distribution of protein numbers was presented for each cluster. (e–f) ECM proteins in cluster 2 (e) and cluster 3 (f) were listed from highest changes to lowest changes. The color of proteins was consistent with the sub-category in pie chart. (g–h) GO analysis of ECM proteins in cluster 2 and cluster 3, including biological process (g) and molecular function (h). Red color represents the ECM proteins expressed highest in MN-ECM in cluster 2 and blue color represents the proteins expressed lowest in MN-ECM in cluster 3. Triplicate independent experiments were performed for each group.