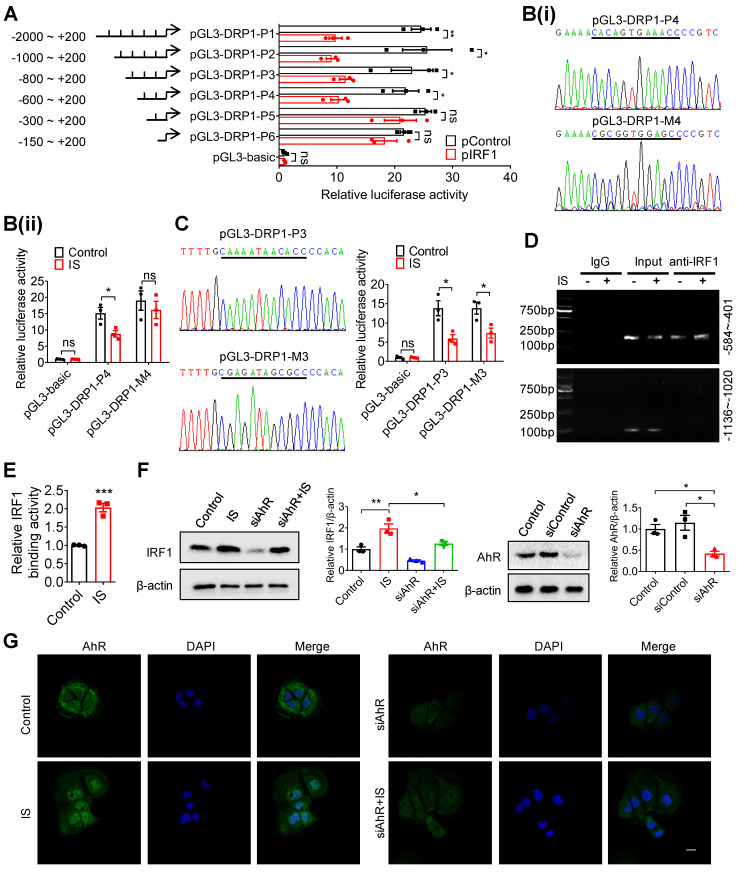
Figure 6.
IRF1 suppresses DRP1 transcription through directly binding to its promoter region. (A) Caco2 cells were co-transfected with pRL-TK vector and pGL3-basic or recombinant plasmids containing various DRP1 promoter regions, in combination with pControl or pIRF1. Then, cells were harvested for dual-luciferase reporter assay. Firefly luciferase activity was normalized against Renilla activity. (B, C) Cells were co-transfected with pRL-TK and pGL3-basic, pGL3-DPR1-P4, pGL3-DPR1-M4, pGL3-DPR1-P3 or pGL3-DPR1-M3 (the mutated IRF1 binding sites were underlined) using Lipofectamine 2000, and treated with control or IS for another 24 hours for dual-luciferase reporter assay. (D) Cells were treated with control or IS for 24 hours and collected for ChIP assay. DNA was amplified using PCR primers (-584 ~ -401 and -1136 ~ -1020). (E) qPCR analysis of IRF1 antibody-immunoprecipitated DNA in (D). (F, G) Cells were transfected with siAhR or siControl and treated with control or IS for another 24 hours. Cells were harvested for Western blot analysis to detect AhR and IRF1 expression (F) and for immunofluorescence confocal microscopy to evaluate the cellular localization of AhR (G). Scale bar, 10 μm. The gray scale of bands was quantified using ImageJ software. Data are shown as mean ± SEM and were analyzed by two-tailed unpaired Student's t test (A-C, E) or one-way ANOVA (F). n=3. ns: no significance. * P<0.05, ** P<0.01, *** P<0.001.