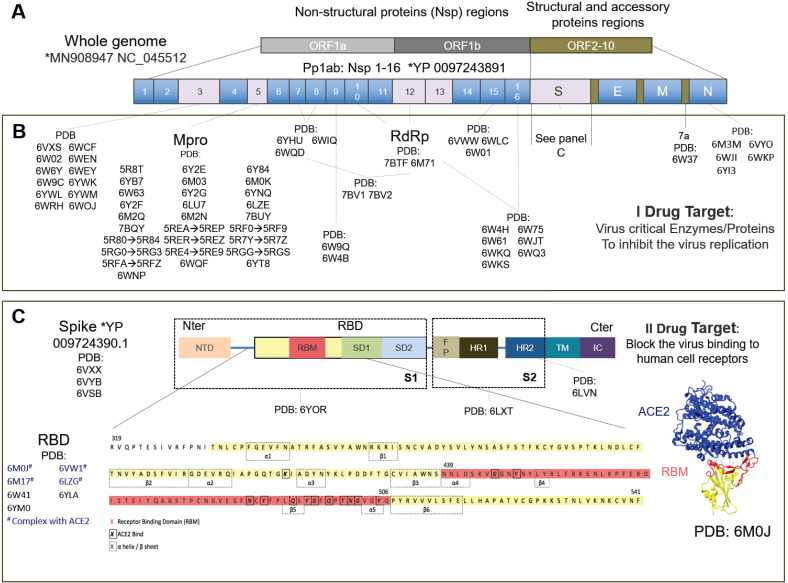
Figure 1.
Drug targets specific to the SARS-CoV-2 virus. A) The SARS-CoV-2 whole genome consists of three main open reading frames (ORF): ORF1a, ORF1b and ORF2-10. The most frequent targets for therapies are reported in gray with their PDB codes. B) Some proteins encoded in ORF1a and ORF1b represent the primary I potential drug target. Their inhibition blocks the viral RNA synthesis and replication. Pp1ab encodes for 16 non-structural proteins (NSPs) including the papain-like protease (PLpro) in the Nsp3region, main protease or C-like protease (Mpro or 3CLpro), ADP ribose phosphatase (ADRP), RNA-dependent RNA polymerase (RdRp) and helicase. ORF2-10 encodes for accessory proteins (e.g. 7a) and the structural and accessory proteins, spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N). C) The spike protein is the most studied secondary II potential drug target. The virus cannot bind the cell receptors if the spike protein is inhibited 14. The spike protein has been studied the most, has been solved by electron microscopy (EM) and consists of: N-ter domain (NTD); receptor binding domain (RBD) consisting of receptor binding motif (RBM), subdomain 1 (SD1) and subdomain 2 (SD2); fusion peptide (FP); heptad repeat 1 (HR1); heptad repeat 2 (HR2); transmembrane region (TM) and intracellular domain (IC). The most studied RBD domain is RBM (in red), the domain mainly involved in host-cell interaction 13. RBM is believed to bind mainly with the ACE2 human cell receptor and, therefore, most of the relevant PDB codes, of which 6M0J, includes the RBD+ACE2 complex. The complete amino acid sequence for RBD is shown at the bottom of the figure. Amino acids corresponding to RBM are in red, the beta-sheet and alpha-helix structures are inside the dotted boxes. The contact residues at the RBD/ACE2 interface 14 are shown in bold squares. *https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/.