Fig. 2.
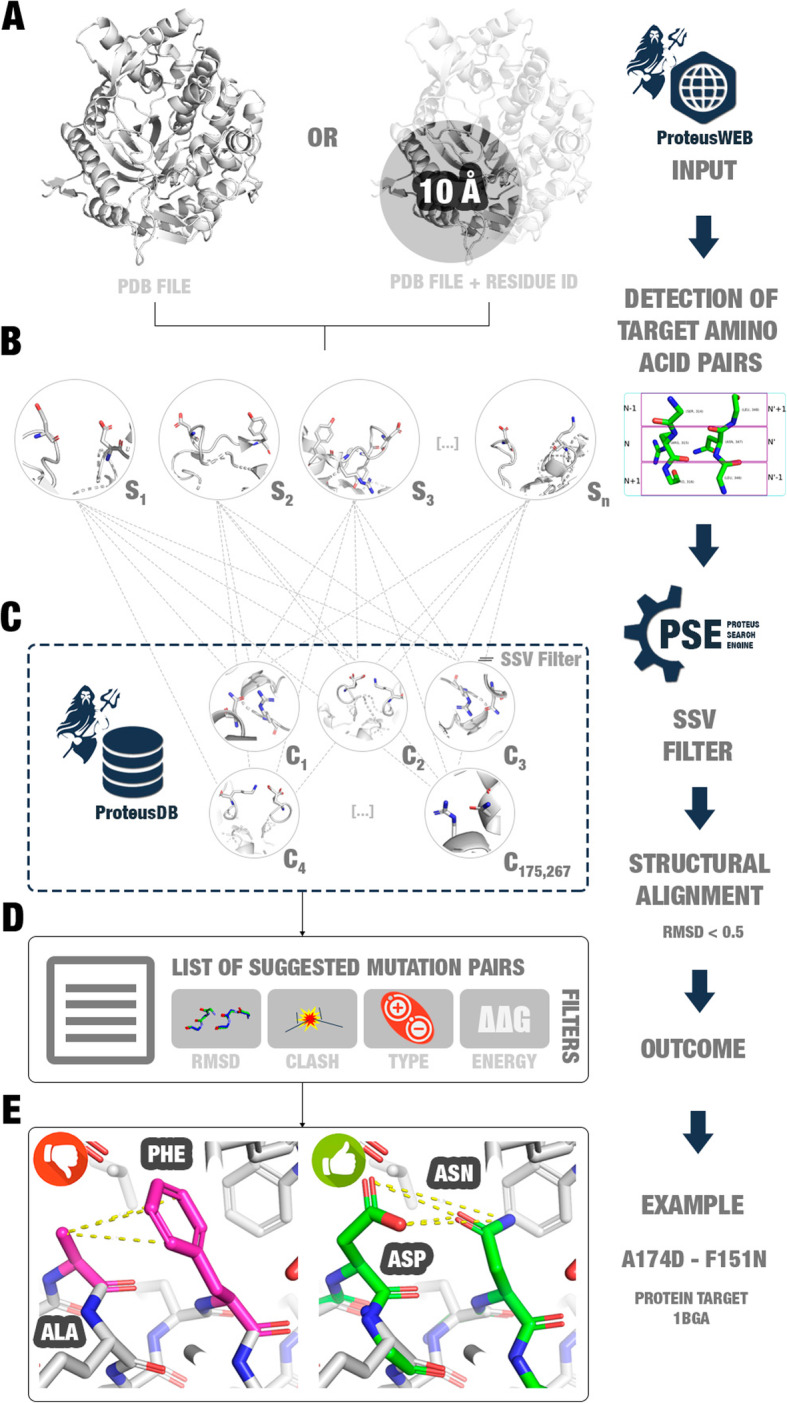
Proteus’ workflow. a ProteusWEB receives as input a PDB file or a region to search for possible beneficial mutations. b ProteusWEB detects target amino acid pairs (alpha carbons at a distance between 3.35–16.40 Å). c Each triad pair containing the target pair is aligned against ProteusDB triad pairs. Only main chain structural alignments between triad pairs from the target and ProteusDB with RMSD < 0.5 Å are selected. d ProteusWEB returns a list of suggested mutation pairs, which can be filtered by users. e Example of a suggested mutation pair for a β-glucosidase enzyme (PDB: 1BGA)
