Figure 1. Genomic Alterations and their Associations with mRNA, Protein, and Phosphoprotein Abundances.
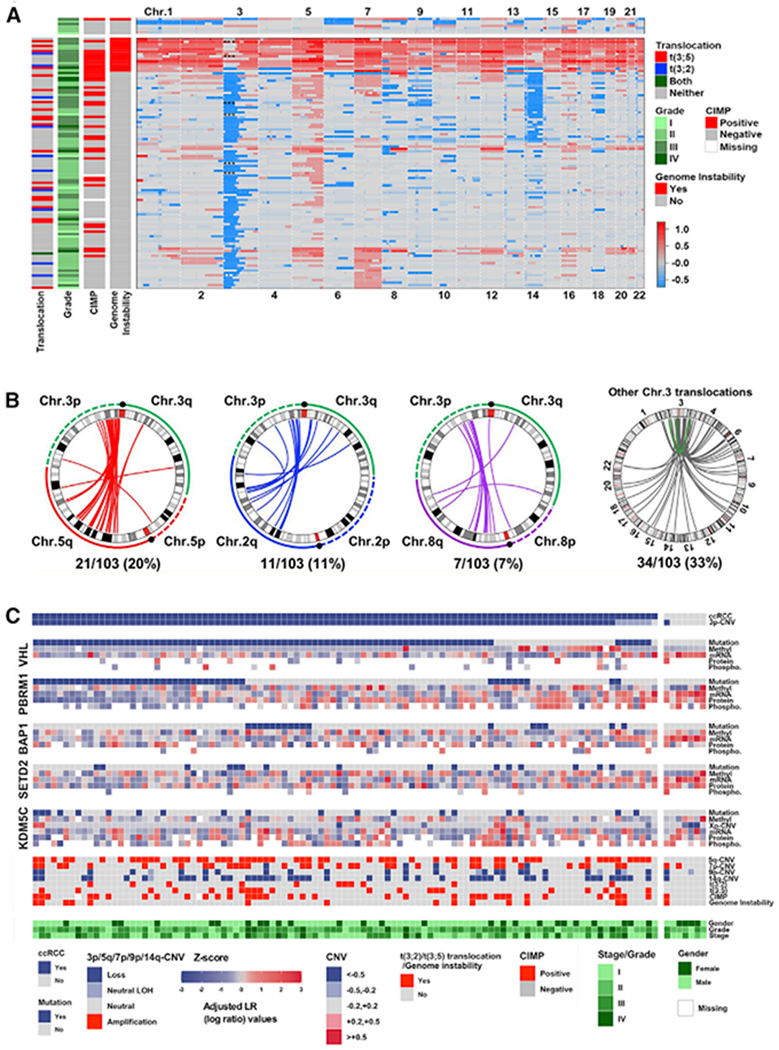
(A) Profiling of absolute copy number estimates observed in the CPTAC cohort. Genomically defined non-ccRCC tumors are above ccRCC tumors; translocation event, grade, CpG island methylator phenotype (CIMP) status, genome instability, and CNV loss/gain are indicated by color coding. ccRCC tumors with evidence of 3p loss of heterozygosity (LOH) are indicated by three asterisks (***).
(B) Circos plots of translocation events involving chromosomes 3 and either chromosomes 5 (red), 2 (blue), 8 (purple) or all other chromosomes (gray), including chromosomal inversion within chromosome 3 (green). Percentage of involved tumors with re-arrangement for each chromosome is annotated below each plot.
(C) Heatmap of multi-omic data for the five key tumor suppressor genes (VHL, PBRM1, BAP1, SETD2, and KDM5C) (n = 103). Tumor samples were ordered by 3p CNV alteration (loss to neutral). Non-ccRCC tumors are separated (right). CNV event, Z score, CNV loss/gain, translocation status, CpG island methylator phenotype (CIMP) status, genome instability, grade, and gender are indicated by color coding (bottom).
