Abstract
Ever present hurdles for the discovery of new drugs for cancer therapy have necessitated the development of the alternative strategy of drug repurposing, the development of old drugs for new therapeutic purposes. This strategy with a cost-effective way offers a rare opportunity for the treatment of human neoplastic disease, facilitating rapid clinical translation. With an increased understanding of the hallmarks of cancer and the development of various data-driven approaches, drug repurposing further promotes the holistic productivity of drug discovery and reasonably focuses on target-defined antineoplastic compounds. The “treasure trove” of non-oncology drugs should not be ignored since they could target not only known but also hitherto unknown vulnerabilities of cancer. Indeed, different from targeted drugs, these old generic drugs, usually used in a multi-target strategy may bring benefit to patients. In this review, aiming to demonstrate the full potential of drug repurposing, we present various promising repurposed non-oncology drugs for clinical cancer management and classify these candidates into their proposed administration for either mono- or drug combination therapy. We also summarize approaches used for drug repurposing and discuss the main barriers to its uptake.
Subject terms: Drug development, Drug development
Introduction
Cancer is one of the leading causes of mortality worldwide.1,2 Opportunities to help reduce the death rate from cancer through the discovery of new drugs are benefiting from the increasing advances in technology and enhanced knowledge of human neoplastic disease.3,4 However, translation of these new drugs into clinical practice has been far slower than expected.5,6 Drug development requires an average of 13 years research. In addition to design and production, it is necessary to examine the efficacy, toxicity, and pharmacokinetic and pharmacodynamic profiles of the drug in cell- and animal-based studies.7,8 Bringing a single new drug from bench to bedside is expensive, with costs of bringing a new chemical entity to market being estimated at ~USD2–3 billion.9,10
A key step in drug development is testing the safety and efficacy in human subjects in clinical trials that normally comprise four phases.11 Phase I clinical trials test the new drug for the first time in a small group of people (e.g., 20–80) to evaluate safety (e.g., to determine a safe dosage range and identify side effects). Phase II clinical trials study intervention in a larger cohort (several hundred) to determine efficacy and to further evaluate drug safety. In phase III studies efficacy is then studied in large groups of trial participants (from several hundred to several thousand) comparing the new intervention to other standard or experimental interventions (or to non-interventional standard care). Phase III studies also monitors adverse effects and collects further information that will allow the intervention to be used safely. Phase IV studies occur after the drug has been marketed. These studies are designed to monitor the effectiveness of the approved intervention in the general population and to collect information about any adverse effects associated with widespread use over longer periods of time. In general, if the drug is found efficacious in Phase III trials, it receives FDA approval. However, only one of every 5000–10,000 prospective anticancer agents receives FDA approval and only 5% of oncology drugs entering Phase I clinical trials are ultimately approved.12,13 Recently, the escalating cost and timeline required for new drug development means that if drug resistance arises, patients with advanced disease may die before alternative treatments become available.14,15
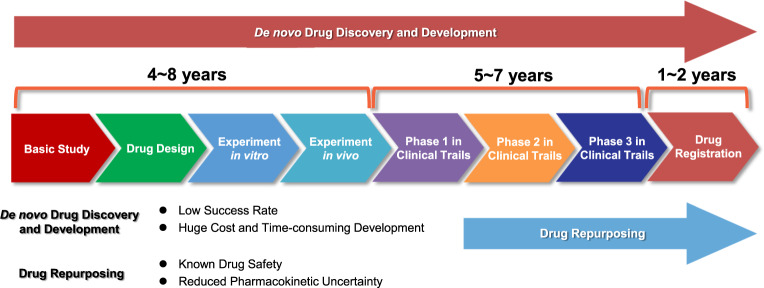
Drug repurposing (alternatively called “new uses for old drugs”) is a strategy for identifying new uses for approved or investigational drugs that are outside the scope of the original medical indication.16,17 Increasingly, researchers and clinicians are considering this strategy to alleviate the dilemma of drug shortage for finding new cancer therapies.18 The major advantage of this approach is that the pharmacokinetic, pharmacodynamic, and toxicity profiles of drugs have been already established in the original preclinical and Phase I studies. These drugs could therefore be rapidly progressed into Phase II and Phase III clinical studies and the associated development cost could be significantly reduced.10,19 Thus, drug repurposing holds the potential to result in a less risky business plan with lower associated development costs, especially if failures of new drugs during research and development (R&D) are factored in (Fig. 1).20,21
Fig. 1.
The estimated time and main steps in de novo drug discovery and development and drug repurposing for cancer therapy. De novo drug discovery and development for cancer therapy takes 10–17 years and comprises basic discovery, drug design, in vitro and in vivo experimentation (including identifying safety and efficacy), clinical trials and finally drug registration into the market. In contrast, drug repurposing for cancer therapy takes only 3–9 years as it can bypass several processes that have been completed for the original indication if the anticancer potential of the candidates is confirmed
In recent years, advanced genomic and proteomic technologies for the assessment of cancer specific biological pathways leading to new drug targets has escalated.22–24 This provides excellent opportunities for drug repurposing.25 Almost all drugs used in human therapy have the potential to address more than one target.22,26 Thus, if the targets of these drugs are highly consistent with cancer, there is a high likelihood that those which share common targets could be therapeutic for other cancer patients.27,28 However, drug repurposing historically has been largely opportunistic and serendipitous.5,29 Indeed, the several successful examples of drug repurposing to date have not involved a precision therapeutic approach.30 Thus metformin, originally an antidiabetic drug, was serendipitously found to be effective in the treatment of various cancers, although the mechanisms for its antineoplastic activity still remain elusive.31–33 Undoubtedly the approach based on oncogene targets, due to its precision and flexibility, aids drug repurposing. However, the remarkable heterogeneity of neoplastic diseases poses an obstacle to such strategies.34–36 It may be more effective to use an understanding of the “the hallmarks of cancer” to mine “the new tricks of the old drug”, rather than blindly pursue similar targets thus missing out on the “metformin-like” success.
In this review, we will mainly focus on the anticancer activity of existing drugs that were not originally intended for cancer therapy to highlight the relevant signaling pathways and discuss the properties of these agents for the reasonable use of medication based on the hallmarks of cancer. Those targeting sustaining proliferative signaling, resisting cell death, deregulating cellular energetics and avoiding immune destruction may be more effective in monotherapy, while those targeting evading growth suppressors, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, genome instability and mutation and tumor-promoting inflammation may be more suitable for drug combination therapy.37,38 We also overview the various approaches that contribute to drug repurposing and discuss the major challenges encountered to date in drug repurposing. Overall, we hope that this article may help researchers and clinicians to get a deeper understanding of drug repurposing based on the ten hallmarks of cancer and help translate old drugs into accepted cancer guidelines.
Non-oncology drugs in drug repurposing
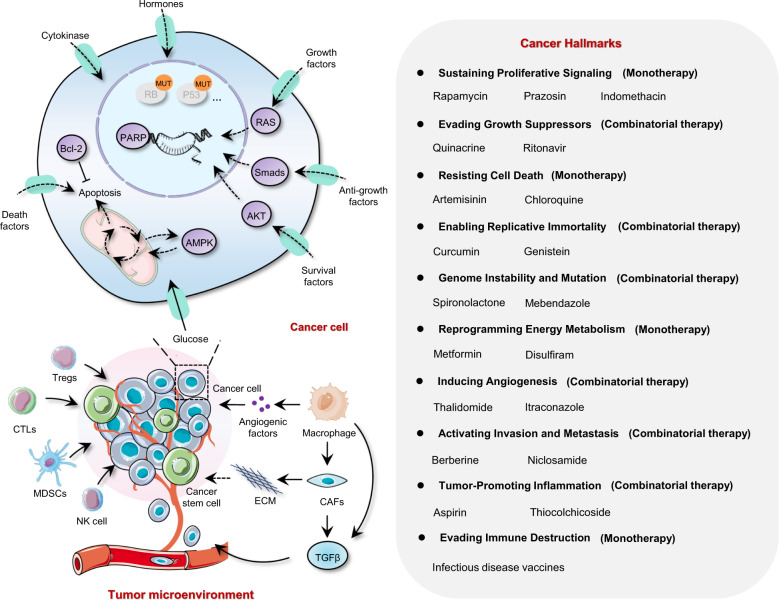
Increasing attention is now being paid to precision medicine, which allows healthcare to be finely tuned to each individual based on molecular profiling instead of the “on drug fits all model” currently used. This has consequentially driven the generation of a biologically compelling lists of genes and proteins based on a systematic analysis of drug discovery for cancer therapy emerging from large-scale multi-omics initiatives.39–41 Drug repurposing can target this to find druggable cancer-associated proteins.28,42,43 However, a major challenge in cancer drug discovery is to maximize the probability that drugs discovered by either biochemical or phenotypic methods will lead to clinical efficacy and improved disease management.8,30,44,45 It should be realized that cell-based models frequently do not accurately represent the true in vivo situation and cancer hallmark-targeting drugs may afford better opportunities for drug repurposing.46–48 Key hallmarks of malignancy are clearly not regulated by a single signaling pathway.37,49 Hence, mono- or multi-hallmark-targeting drugs have advantages since they may target several supporting pathways pharmacologically, thereby partially avoiding the progression of adaptive resistance.37,50 We now summarize non-oncology compounds used for cancer therapy that target the hallmarks of cancer, and distinguish between those agents suitable for monotherapy or as drug combinations (Fig. 2).
Fig. 2.
Identification of drug candidates targeting the hallmarks of the cancer cell using drug repurposing enabled by recapitulative signaling networks. The complex signaling interactions contributing to the hallmarks of cancer cells can be orchestrated, rationalizing the complexities of neoplastic disease. Drug candidates interfering with cancer capabilities are shown. CAFs cancer-associated fibroblasts, CTLs cytotoxic T lymphocytes, ECM extracellular matrix, MDSCs myeloid-derived suppressor cells, NK cells natural killer cells, Tregs regulatory T cells
Non-oncology drugs suitable for cancer monotherapy
Many newly identified non-oncology drugs repurposed for cancer therapy act by inhibiting proliferation and inducing cell death as indicated by a large body of data from in vitro and in vivo experiments or clinical trials.51–53 In addition, these drugs, which have previously been used for other indications, already have reliable drug safety data and are often inexpensive (especially if they are available as generics).54 Some of these drugs are summarized below.
Non-oncology drugs which work by inhibiting proliferative signaling
A fundamental trait of cancer cells is their ability to maintain chronic proliferation. Cancer cells can therefore master their own destiny, becoming self-sufficient with respect to growth signaling.37,55 Based on an understanding of the underlying cancer biology, various molecular targeting agents, like receptor tyrosine kinase inhibitors, have been developed with excellent success as cancer therapeutics. However, relatively few drugs targeting these pathways have been discovered by drug repurposing.56,57 Remarkably, cancer cells may also circumvent pathways that need ligand-mediated receptor induction to sustain proliferative signal, instead relying heavily on three important pro-survival signaling pathways: PI3K/AKT, mTOR, and MAPK/ERK.58,59 An ever-increasing number of non-oncology drugs have been repurposed to treat cancer by effective inhibition of these pathways.
Rapamycin
Rapamycin allosterically inhibits mTORC1 by binding to the FRB domain of mTOR.60 It was approved in 1999 as an immunosuppressant for preventing kidney transplant rejection due to effective suppression of IL-2-mediated T cell proliferation.61 In 2002, rapamycin also received clinical approval as an anti-restenosis agent for inhibiting the growth of vascular smooth muscle.62,63 However, it was repurposed with the anticancer potential in recent years due to an improved understanding of the role of mTOR and associated signaling networks in cancer.60,64–66 Earlier studies reported that the number of leukemic progenitor cells in patients with acute myeloid leukemia was decreased after rapamycin treatment.67,68 In addition, rapamycin also showed effective anticancer activation in drug resistant chronic myelogenous leukemia with only mild side effects in most patients.69,70 However, more recent studies are highlighting some defects of rapamycin for single-agent therapy. First, the inhibition of mTOR1 induces negative feedback regulation and activation of PI3K-AKT signaling causing survival of cancer cells.71 Second, proteins downstream of mTOR such as 4EBP1, a translation repressor protein can be reactivated to drive the proliferation of cancer cells under long-term rapamycin treatment.72,73 In addition, rapamycin has limited influence on the activation of mTORC2, which plays an active role in tumorigenesis as an important part of the PI3K–mTORC2–AKT axis.60,74 However, some combinatorial strategies of mTOR inhibitors in the clinic have proved beneficial. For instance, the combination of insulin like growth factor 1 receptor (IGF1R) inhibitors with semisynthetic rapamycin analogs inhibits the proliferation of breast and prostate cancers and myelomas.60,75–77 Similar encouraging results come from other growth factor receptor antagonists, including the epidermal growth factor receptor (EGFR).78 Recently, an AKT inhibitor, perifosine, was used to treat multiple myeloma cells and potentiate the anticancer activity by combination with nanoparticle bound rapamycin.79 Furthermore, several clinical trials, in which a RAF pathway inhibitor, sorafenib, or histone deacetylase inhibitor, vorinostat, combined with rapamycin, are underway.80,81 It is worth noting that temsirolimus, a rapamycin analog with better solubility and specificity, was approved by the FDA and the European Medicines Agency for the treatment of renal cell carcinoma in 2007.82
Prazosin
Prazosin was approved for the clinical treatment of hypertension over 40 years ago.83,84 The current use of prazosin has been extended to treat multiple clinical indications, such as benign prostatic hyperplasia, Raynaud’s disease and congestive heart failure.85–87 Pharmacologically, prazosin is known to be a non-selective inhibitor of α1- adrenergic receptor and can selectively inhibit α2B-adrenergic receptor.87–89 Prazosin, showing potential anticancer effects, has been recommended for the treatment of pheochromocytoma.90,91 In addition, Desiniotis et al. reported that doxazosin and terazosin, quinazoline-based agents, induced apoptosis by activating the TGF-β signaling pathway rather than adrenergic receptor associated pathways in prostate cancer, suggesting prazosin related drugs, with a well-established and documented clinical record, have potential for cancer therapy by off-target associated drug repurposing.92 A recent study indicated that prazosin also induced growth inhibition in a concentration-dependent manner in patient-derived glioblastoma-initiating cells (GICs). Interestingly, there was an off-target mechanism of prazosin in GIC treatment, evidenced by preferential toxicity among other quinazoline-related antagonists of α-adrenergic receptor and activation of the MAPK/ERK signaling pathway followed α-adrenergic receptor blockage. They found that prazosin inhibited GIC proliferation by inhibiting the PKCδ-dependent AKT signaling pathway, but rarely influenced AKT-dependent growth of neural stem cells due to a paucity of PKCδ compared with GICs.93 Taken together, these studies imply drug repurposing of prazosin holds promise for clinical cancer treatment.
Indomethacin
Indomethacin is a non-steroidal anti-inflammatory drug (NSAID) used for rheumatic disease treatment regimens due to its antinociceptive action.94,95 Interestingly, multiple clinical studies have found patients undergoing long term NSAID treatment have a lower risk of developing cancers.96,97 In addition, there are an increasing number of reports indicating the antineoplastic effect of indomethacin and indomethacin-based prodrugs against a wide range of cancers by the inhibition of Cox-1/2-dependent angiogenesis.98,99 Furthermore, indomethacin has also been shown to reduce the migration and invasion of cancer cells by interfering with calcium-associated signaling and formation of focal complexes.100 Several recent studies have uncovered a Cox-independent mechanism of action for the antiproliferative effect of indomethacin, as evidenced by the inhibition of cell proliferation in indomethacin treated colorectal cancer cell lines that do not express Cox-1/2.101–103 Recently, Lin et al. suggested that the antiproliferative effect of indomethacin resulted from the inhibition of MAPK-related pathways.104 They executed drug repurposing using a library of existing drugs with computational screening. Using binding to the phosphotyrosine binding (PTB) domain of adapter protein Shc (ShcPTB) as a readout, indomethacin was confirmed to have a direct interaction with ShcPTB. They further showed that indomethacin competes with activated EGFR by binding to ShcPTB without disruption of the ERK-binding site, resulting in a failure of EGFR to recruit Shc to induce aberrant signaling due to the release of ERK. In addition to targeting MAPK pathways, indomethacin also inhibits the proliferation of cancer cells by impairment of PKCζ-p38-DRP1 axis-dependent mitochondrial dynamics or downregulation of Wnt/β-catenin signaling.105 Various NSAIDs including indomethacin, have now been proposed as anticancer chemo-preventive drugs.106–109 With further understanding of the detailed mechanisms involved in their antineoplastic effect, the clinical application of indomethacin shows much promise.
Non-oncology drugs which work by inducing cell death
Apoptosis, which causes cell death once the cell is damaged or faces various physiologic stresses, is a natural barrier for tumorigenesis.110 In cancer, deregulated apoptotic signaling, particularly activation of antiapoptotic systems, allows cancer cells to escape this program resulting in uncontrolled proliferation, tumor survival, therapeutic resistance and cancer recurrence.111 However, even if tumor cells evolve multiple strategies to antagonize or circumvent apoptosis, other programmed cell death modalities, like lethal-autophagy and ferroptosis, could provide alternative strategies for cancer treatment.37,55,112 Increasingly, non-oncology drugs are being repurposed for the treatment of apoptotic resistant cancers by triggering cell death mechanisms.
Artemisinin
Artemisinin (ARS) is the active ingredient of artemisia annua L and is used to treat malaria, the world's most prevalent disease affecting over 600 million people each year.113 Artemisinin and its derivatives have caught worldwide attention over the past years, and artemisinin-based therapies of malaria are now the established clinical standard.114–116 The main mechanism by which artemisinin kills plasmodia is by blocking conversion of hemoglobin to the non-toxic hemozoin in malaria parasites, resulting in accumulation of reactive oxygen species (ROS) from heme-iron.117–120
Interestingly, Youyou Tu, who was honored with the Nobel Prize in Physiology or Medicine for first extracting dihydroartemisinin (DHA), found that patients with lupus erythematosus-related nephritis might benefit from DHA due to inhibition of the NF-κB signaling pathway.121 In addition, increasing studies reported ARS-related drugs had an unexpected therapeutic effect against viruses (human cytomegalovirus), schistosomiasis and trypanosomiasis.122–124 Recent results indicated ARS and its derivatives also had anticancer activity attributed to inducing non-apoptotic programmed cell death.54,125,126 For instance, artesunate (ART)-based agents stimulated ROS generation and promoted the lysosome degradation of ferritin instead of autophagic degradation, resulting in system Xc-/GPX4 axis-mediated ferroptosis in cancer cells.127 Similarly, Chen et al. found that DHA sensitized cancer cells to ferroptosis by autophagic independent degradation of ferritin, inducing free iron accumulation and subsequently iron homeostasis dysregulation due to binding to the iron-responsive element sequences of iron-regulatory proteins.128 In contrast, Du et al. reported DHA-induced cell death associated with autophagic-dependent degradation of ferritin (ferritinophagy) in acute myeloid leukemia cells.129 In addition, several targets involved in autophagy of ARS and its derivatives were identified.54 Dihydroartemisinin-37 induced autophagic cell death by activating high mobility group box 1 and releasing Beclin 1 from Bcl-2.130 Other types of nonapoptotic programmed cell death, including oncosis (ischemic cell death) and anoikis (anchorage-dependent cell death), have also been observed in cancer cells following ARS-based treatment.131–133 In recent clinical trials, DHA treatment for advanced cervical carcinoma patients appeared encouraging as clinical symptoms of these patients were relieved and thus survival increased.134 In addition, artesunate administered orally in patients with colorectal cancer showed anticancer effects and was generally well tolerated.135 23 metastatic breast cancer patients who were also enrolled to receive ARS-type drugs treatment showed beneficial effects.136 Taken together, drug repurposing of ARS and its derivatives, like ART and DHA, offers an opportunity for treating intractable and antiapoptotic cancer by inducing alternative programmed cell death. Further Phase II, Phase III clinical trials will further confirm the suitability of ARSs in clinical oncology.
Chloroquine
Chloroquine (CQ) and its derivative hydroxychloroquine (HCQ), which are antimalarial agents in common clinical use, also have therapeutic effect on rheumatoid arthritis and discoid and systemic lupus erythematosus.137,138 CQ or HCQ, the only FDA-approved autophagy flux inhibitors, have been used to treated pancreatic and other cancers.139,140 Mechanically, CQ or HCQ blocked autophagosome-lysosome fusion through blocking STX17 incorporation by LC3-positive autophagosomes, or impairment of Golgi and endosomal functions, rather than decreasing lysosomal acidity.141 Recently, a novel CQ derivative DC661 was reported to have a deacidifying function toward lysosome and had distinct advantages in inhibition of autophagy flux compared with HCQ.142 DC661, and other CQ-based drugs, can target and inactivate palmitoyl-protein thioesterase 1 (PPT1), which mediates depalmitoylation to stabilize the v-ATPase subunits located in lysosome. The v-ATPase complex not only maintains the acidity of lysosome for catabolism, but contributes to the localization and subsequent activation of mTOR. In addition, high expression of PPT1 in a variety of cancers was identified using the The Cancer Genome Atlas (TCGA) database, and indicated poor survival.142 These findings further confirm the specific targets of CQ-based drugs and broaden the clinical application of CQ-based drugs to patients with tumors exhibiting PPT1-high expression.142 CQ-related clinical cancer trials started as early as 1998 when CQ was used for treating glioblastoma multiforme patients showing enhanced patient survival.143 Subsequently, CQ or HCQ was used alone or in combination with standard treatments for multiple myeloma, lung cancer, pancreatic cancer, and sarcoma.144–148 Drug repurposing of CQ shows considerable promise for autophagy-dependent malignant tumors, including pancreatic cancer and drug resistant cancer mediated by protective autophagy.
Non-oncology drugs which work by regulation of cellular metabolism
Reprogramming energy metabolism is ubiquitous in cancer cells, and primarily supports malignancy by sustaining key hallmarks of cancer, including uncontrolled cell proliferation, evading growth suppressors, and resisting cell death.37,149 Cancer cells exhibit anomalous energy metabolism, termed aerobic glycolysis, which generates the glycolytic intermediates to facilitate the production of macromolecules and organelles by various biosynthetic pathways. Accordingly, increased aerobic glycolysis significantly enhances glucose uptake and utilization in order to provide essential components required for cell assembly regardless of relatively poor efficiency for ATP generation.150,151 In general, reprogramming energy metabolism as a proliferation-inducing phenotype fuels chronic and abnormal cell growth and division.152,153 Notably, changes in metabolic patterns may lead to hyper-activation of certain metabolic enzymes that detoxify endogenous xenobiotics derived from abnormal energy metabolism in cancer cells, resulting in the malignant phenotype including resisting cell death and drug resistance.154,155 Repurposing existing drugs to target cellular metabolism for cancer treatment is therefore a useful strategy.
Metformin
Metformin was approved by the FDA in 1994 for treating obese type 2 diabetes.156,157 Generally, patients with diabetes are more prone to several types of cancer, probably due to chronic and increased glycemia contributing in part to tumor development.158 A number of studies have shown that diabetes, especially type II diabetes, is closely related to the development of pancreatic, bladder, colorectal cancer and non-Hodgkin’s lymphoma.159 Long-term metformin treatment has been shown to substantially lower the risk of breast cancer in women with type 2 diabetes160 and to date, more than 100 ongoing clinical trials are investigating the anticancer activity of metformin, using those doses typically used for diabetes, for cancer therapy.161 Mechanically, metformin can lower elevated levels of insulin, which activates the PI3K-mTOR signaling pathway, leading to proliferative inhibition of those cancers with insulin receptor expression.161,162 Furthermore, metformin has been reported to activate AMPK, a key energy sensor in cellular metabolism, to inhibit cancer cell growth by the negative regulation of mTOR involved in tumor survival.163 In contrast, studies have indicated metformin inhibition of mTOR signaling dependent on Ras-related GTPase but independent of AMPK.164 Wu et al. further showed that metformin protected tumor suppressor TET2 phosphorylation at serine 99 by activating AMPK, thereby avoiding the destabilization of TET2 and dysregulation of 5-hydroxymethylcytosine. Furthermore, metformin has been shown to induce energetic stress by partially inhibiting oxidative phosphorylation, resulting in repression of some high-energy consumption processes, such as mRNA translation and biomacromolecule synthesis, ultimately contributing to cytostatic or cytotoxic effects.165 Recently, Liu et al. reported that metformin mediated tumor cell-intrinsic mitochondrial metabolism in ovarian cancer.166 Subsequently, a mitochondrial metabolism-related protein, BTB and CNC homology1 (BACH1), was found to increase expression in triple-negative breast cancer, leading to poor glucose utilization and down-regulated transcription of electron transport chain genes. This study showed that targeting BACH1 sensitized breast cancer to metformin treatment.167 Taken together, metformin used in cancer therapy is one of the most successful cases of drug repurposing and several clinical trials, which have advanced to Phase III and Phase IV, are currently investigating the therapeutic potential of metformin in oral, prostate, breast, endometrial, and pancreatic cancers.168–171
Disulfiram
Disulfiram was originally used in rubber vulcanization, but subsequently has been used for more than 60 years as an alcohol-aversion drug to treat alcohol abuse.172,173 Mechanistically, disulfiram inhibits the activation of acetaldehyde dehydrogenase (ALDH), triggering severe nausea and vomiting symptoms contributing to abstinence.174,175 In the past years, disulfiram has attracted increasing attention for its anticancer activity both as monotherapy and combinational therapies.176 Notably, numerous studies have reported on mechanisms of action of disulfiram that are closely associated with ALDH-related processes of cellular metabolism. For example, Tacconi et al. identified disulfiram-mediated acetaldehyde metabolism as a potential therapeutic target for BRCA1/2-deficient cancer cells, resulting from endogenous acetaldehyde-induced DNA damage.177 Furthermore, Choi et al. reported that disulfiram reduced the metabolism of ALDH-positive atypical teratoid/rhabdoid tumor cells. They further showed that the disulfiram-induced change of the nicotinamide adenine dinucleotide ratio of NAD+/NADH mediated the function of some NAD+-dependent proteins, like SIRT1, which controlled various important processes in cancer cells including apoptosis, cell differentiation and metabolism.178 Disulfiram can also mediate formaldehyde metabolism by the blockage of formaldehyde oxidation due to inhibition of ALDH, potentially contributing to the induction of apoptosis in various cancer cells.179,180 ALDH has also been reported to play a key role in maintaining the stemness of cancer cells, therefore explaining the anticancer effect of disulfiram in stem-like cancer cells.181–183 In addition to ALDH-related processes, disulfiram regulates copper- or zinc-dependent metabolism processes in cancer cells.84,184 More and more evidence is indicating that disulfiram-mediated oxidative metabolism partially contributes to its anticancer activity.185,186 Notably, clinical trials of disulfiram are ongoing, or have already been completed, for treating patients with melanoma, glioblastoma, breast, prostate, and non-small cell lung cancers (NCT02101008) (NCT01118741) (NCT00312819) (NCT01907165).
Non-oncology drugs which work by activation of antitumor immunity
Some cancers, especially virus-induced cancers, can avoid immune surveillance or limit immunological killing by somehow regulating both the innate and adaptive immune systems, which act as an effective barrier to inhibit tumorigenesis and development, to evade eradication.37,187 Indeed, increased tumor incidence and cancer development partially attribute to certain deficiencies of immunocytes that include natural killer (NK) cells, CD8+ cytotoxic T lymphocytes (CTLs), and CD4+ Th1 helper T cells.37,188,189 Mechanistically, cancer cells may disable the regulators or effectors of the immune system to evade immune destruction.190 An increasing body of evidence suggests that antitumor immunity has a favorable potential to eradicate cancer cells, and is currently revolutionizing cancer care.191 However, drugs like immune-checkpoint inhibitors for cancer immunotherapy are not effective in all cases, and would benefit from drug repurposing.
Infectious disease vaccines
Vaccines act by stimulating the body’s defense mechanisms against infection to produce the corresponding antibodies.192 Recently, mounting evidence has indicated that intratumoral administration of certain infectious disease vaccines exert an anticancer effect by largely eliciting or potentiating tumor immune responses usually manifested as recruitment and activation of immunocytes.193 For example, Shekarian et al. reported that rotavirus vaccines activated NF-κΒ and type I interferon pathways in a retinoic acid induced gene 1-mediated manner instead of a Toll-like receptor-dependent manner.194 Furthermore, rotavirus vaccines elicited tumor immune responses by enhancing the infiltration of CTLs, NK, and CD4+ Th1 helper T cells while the intratumoral administration induced immunogenic cell death, evidenced by ATP release. They also identified the senitization of rotavirus vaccines to overcome anti-CTLA-4 cancer immunotherapy resistance.194 In addition, a prophylactic vaccine of yellow fever (live 17D) was shown to exert anticancer activity against cancer cells by the regulation of CTLs and reduction of Tregs.195 Interestingly, a previous case–control study reported that anti-influenza vaccines could improve the protective efficacy against cutaneous melanoma, suggesting immune-mediated antitumor activity.196 To date, there have been more than 200 clinical trials that utilize vaccines for cancer therapy and an increasing amount of these have advanced into Phase III and Phase IV clinical trials (https://clinicaltrials.gov/). Taken together, attenuated viral vaccines for intratumoral immunotherapy from drug repurposing play a key role in exerting immunostimulatory and oncolytic properties in tumor immunotherapy. Furthermore, it can accelerate the clinical development based on documented safety records. Notably, these approved and marketed agents have shown promising for treating infants and children with cancers.193
Non-oncology drugs suitable for drug combination therapy
In addition to those mentioned above, other compounds may be neglected by researchers for drug repurposing screening due to their low anticancer activity at known tolerated plasma drug doses described in previous indications, hindering their application and development. Fortunately, drugs that may be effective at higher dosage still have a chance to be realized. Indeed, drug combination therapy may be able to utilize such effects and repurpose these drugs, producing a synergistic effect by targeting alternative signaling pathways associated with certain cancer hallmarks, therefore sensitizing cancer cells to other cytotoxic agents. In addition, drug combination therapy may result in reduced doses of the individual agents.197 Notably, neoplastic disease has been identified with multifactor and polygenic pathologies, suggesting the requirement to target cancer-related signaling networks using drug combination therapy.198 Indeed, drug combinations of two or more compounds is now common practice in the clinic utilizing their diverse mechanisms of action.199,200 These drugs will be summarized below.
Non-oncology drugs which work by reactivating growth suppressors
Cancer cells can evade mechanisms mediated by the action of tumor suppressor genes.37,55 Notably, RB (retinoblastoma-associated) and p53 proteins are two main tumor suppressors, operating in various ways to modulate cells to maintain homeostasis.201–203 Defects in function of the RB pathway result in persistent cell proliferation due to dysregulation of gatekeepers of cell-cycle progression.204 By contrast, p53 senses intracellular stress, such as energy stress, genotoxic stress, and oxidative stress, preventing further cell-cycle progression until homeostasis returns to normal. In addition, p53 can induce apoptosis when irreparable damage occurs.205,206 Accumulating evidence indicates a lack of crucial tumor suppressors (including, but not limited, to RB and p53) stimulating progression to neoplasia.201,207,208 More and more non-oncology drugs are being repurposed to target cancer cells evading growth suppressors.
Quinacrine
Quinacrine is an antimalarial drug discovered in the 1920s.209 It is used to treat giardiasis as an antimicrobial, rheumatoid arthritis, or systemic lupus erythematous as an anti-inflammatory and pneumothorax as a pleural sclerosing agent.210–212 In addition, the clinical evaluation of quinacrine for treatment of Creutzfeldt–Jakob disease is in progress.213–215 Quinacrine has a convincing drug safety record for long-term clinical use compatible with drug repurposing for cancer treatment.215 Indeed, previous studies have indicated quinacrine brings benefits for the treatment of multiple cancers, and its anticancer effects are mediated by p53 activation. Using small molecule screening on renal cell carcinoma cells, quinacrine was shown to induce p53 expression.216 In addition, data indicates the cytotoxicity of quinacrine corresponds with increased p53 levels. Subsequent studies suggested that quinacrine-induced p53 expression was possibly mediated by the Facilitates Chromatin Transcription (FACT) protein complex, which is trapped onto the chromatin thereby inducing CK2-mediated phosphorylation of p53.217,218 In contrast a recent study reports the inconsistent result that p53 knockdown enhances the quinacrine effect in MCF-7 cells compared with controls.219 Regardless, these studies indicate that quinacrine cytotoxicity in cancer cells depends, at least partially, on the p53 status.220,221 Reports of quinacrine acting as an anticancer agent in clinical trials are increasing.222 For instance, the scientists of Fox Chase Cancer Center currently combined quinacrine with capecitabine for treating colorectal adenocarcinoma in a Phase I, Phase II clinical trial (NCT01844076). Quinacrine has also been combined with erlotinib for the treatment of recurrent or late-stage non-small cell lung cancer in a Phase I clinical trial (NCT01839955). Collectively, quinacrine as an anticancer agent has major potential for cancer therapy and the mechanism may be closely related to activation of p53, a key growth suppressor dysregulated in multiple cancers.222
Ritonavir
Ritonavir is a protease inhibitor widely used in highly active anti-retroviral therapy (HAART) to treat human immunodeficiency virus (HIV) infection.223 HAART, in which protease or integrase inhibitors are combined with reverse transcriptase inhibitors, can substantially slow progression of the AIDS virus rendering it manageable.224–226 Interestingly, use of HAART can also result in a reduction in tumor incidence, as evidenced by a Swiss HIV cohort study that reported a decreased incidence of Kaposi sarcoma with HAART treatment.227,228 Other research groups subsequently reported similar conclusion on the potential anticancer effect of HAART.229–231 In fact, ritonavir as a key component of HAART had already been shown to have anticancer activity in several cancers by inducing apoptosis.232 The underlying mechanisms of ritonavir in cancers is controversial but appears to be closely related to key growth suppressors. Gaedicke et al. observed the anticancer activities of ritonavir and accumulation of p53 in ritonavir-treated cancer cells.233 A previous study indicated that ritonavir induced cell death of ovarian cancer. Downregulation of CDKs, which inactivate RB through phosphorylation, was observed in the gene profiles of ritonavir treated cells, resulting in RB dephosphorylation and subsequent cell cycle arrest and apoptosis.234 A similar outcome of ritonavir-induced G0/G1 arrest was observed in lung adenocarcinoma, associated with downregulation of RB phosphorylation.235 Batchu et al. also suggested that ritonavir induced apoptosis in pancreatic cancer, attributing this to inhibition of AKT pathways mediated RB activation and formation of the E2F–1/RB complex.236 In a recent phase II study clinical trial, ritonavir combined with lopinavir was used for treating patients with high-grade glioma. However, compared with standard therapeutic regimes, a patient with complete remission showed no improvement in progression-free survival. The authors proposed that the blood brain barrier probably played a key role in the limited efficacy.237 Ritonavir as a clinically approved HIV drug has potential for adjuvant therapy of cancer by drug repositioning, although some challenges ahead remain.
Non-oncology drugs which work by interfering with replication
In the normal cell lineages, the cell growth and division cycle are controlled by senescence and crisis/apoptosis.238,239 In cancer cells, the specialized DNA polymerase, telomerase, which is related to telomere maintenance, is expressed at high levels to counteract this.240–242 In addition, TERT, a catalytic subunit of the enzyme telomerase acts as a cofactor to amplify Wnt signaling pathways causing cancer cells to revert to a pre-differentiated stem-like phenotype, eventually exhibiting unlimited replicative potential and substantial stress adaptability.243–245 Notably, the Hippo signaling pathways, which control organ size and are dysregulated in cancer, orchestrate Wnt signaling pathways via reciprocal crosstalk with each other by a series of mechanisms.246–248 Accordingly, repurposing of drugs which target telomerase to mediate canonical or noncanonical function (influencing Wnt/Hippo signaling pathways), is an actionable and available strategy for improving efficacy of cancer therapeutics.
Curcumin
Turmeric, belonging to the ginger family that is widely used in the Asian region as a cooking spice has been used to treat dermatological diseases.249,250 Curcumin, the major active ingredient of turmeric, possesses antioxidant, anti-inflammatory, and even anticancer properties.251,252 Recently, the influence of curcumin on telomerase has received widespread attention. It has been proposed as a telomerase inhibitor with substantial potential for cancer therapy by drug repurposing. Curcumin has been reported to down-regulate hTERT at the level of transcription to mediate telomerase activity in breast cancer cells.253–255 In addition, another study indicated that curcumin inhibited telomerase activity by induction of the dissociation of hTERT from p23, leading to cytoplasmic retention of hTERT and subsequent HSP90-mediated proteasome degradation.256 Aravindan et al. showed that curcumin profoundly inhibited telomerase activity by repressing NF-κB binding to the promoter region of the TERT gene following treatment of human neuroblastoma cells with ionizing radiation.257 Similarly, a synthetic curcumin analog was shown to affect cancer stemness and telomerase in colorectal cancer. The drug deactivated STAT3 and NF-κB so that the interaction with the hTERT promoter was decreased, resulting in the inhibition of non-canonical functions of telomerase contributing to stemness.258 Further studies have confirmed the effects of curcumin on the Wnt/β-catenin and Hippo/YAP signaling pathways, inhibiting stemness or proliferation in multiple cancers.259–265 Although limited bioavailability and stability hamper the application of curcumin in clinical cancer therapy,266–268 curcumin has been used for treating patients with pancreatic cancer in clinical trials due to its anticancer activity and safety profile.269,270 Drug repurposing of curcumin could prove beneficial for clinical translation using highly bioavailable forms of curcumin (e.g., micro- and nano-formulations of curcumin with greatly enhanced absorption) to target telomerase.
Genistein
Genistein is an isoflavone found in legumes offering low cost, safe, and dietary accessibility treatment for various pathologies including menopause, osteoporosis, and obesity.271–273 Notably, genistein has been used as a phytoestrogen mimic of 17β-estradiol to treat breast cancer.274 Subsequently, genistein has been shown to inhibit telomerase activity and related onco-signaling pathways giving potential for treatment of multiple cancers.275,276 It has been reported to inhibit transcriptional activation of hTERT by partially attenuating c-Myc in prostate cancer cells.277 Genistein inhibits hTERT nuclear translocation posttranslationally, as evidenced by the downregulation of hTERT phosphorylation mediated by AKT in prostate cancer cells.278 Genistein has also been reported to inhibit glioblastoma, head and neck and other cancers in a telomerase-dependent manner.279–281 In addition, Li et al. discovered genistein-induced telomerase repression due to crosstalk between genetic and epigenetic mechanisms. They indicated that genistein decreased the expression of DNA methyltransferases DNMT1, DNMT3a, and DNMT3b, leading to the hypomethylation of the E2F-1 (repressor) recognition site in hTERT and increased binding of E2F-1 and hTERT. Genistein-induced hTERT reduction has been attributed to increases of the inactive chromatin marker trimethyl-H3K9 and decreases of the active marker dimethyl-H3K4 to the hTERT promoter.282 Recently, genistein was reported to inhibit Wnt/β-catenin signaling by regulation of related genes, microRNA, DNA methylation and histone modification, inducing inhibition of proliferation or apoptosis in various cancers.283–285 Furthermore, genistein also inhibited stemness, which is closely related to the developmental pathways including Wnt/β-catenin and GDF15, in renal and other cancers.283,286,287 Clinical trials of genistein in cancer therapy have been conducted, including in breast cancer (NCT00244933) (NCT00290758), in breast or endometrial cancer (NCT00099008), in bladder cancer (NCT00118040), and in prostate cancer (NCT00005827). However, clinical applications of genistein are still limited due to the poor solubility and bioavailability regardless of its potential anticancer activity by drug repurposing.288 Interdisciplinary studies, leading to improved formulation and drug delivery may extend the potential of genistein for clinical anticancer treatments.
Non-oncology drugs which work by decreasing angiogenesis
Tumor cells stimulate angiogenesis to generate neovasculature, an important mechanism by which tumors obtain nutrients and evacuate waste products. By contrast, benign neoplasias are relatively dormant, which is attributed to insufficient blood supply.289,290 However, when dormant tumor cells are activated by angiogenesis, secreted growth factors induce endothelial cells to bud, leading to a chemotactic response and an angiogenic switch.291,292 Antiangiogenesis therapies rely on reducing blood vessel density resulting in tumor starvation.289,293 However, antiangiogenic agents are still deficient and usually stimulate cancer to develop stress resistance states.294 Accordingly, the identification of more effective drugs with antiangiogenic activity by drug repurposing is important for improving cancer therapy.
Thalidomide
Thalidomide is a glutamic acid derivative originally proposed as a sedative in 1957 and was later used as an antiemetic for treating the symptoms of morning sickness in pregnant women. With an improved understanding of the mechanisms of action of thalidomide, it was found to have immunomodulatory, anti-inflammatory, antiangiogenesis and cell proliferation inhibitory properties suitable for treating various diseases.295–298 Notably, thalidomide in combination with dexamethasone was approved by the FDA for treating multiple myeloma in 2006.299 Further studies have indicated thalidomide has marked anticancer activity in various cancers by mediating angiogenesis.300 Nowadays, thalidomide is widely recognized as an antiangiogenic agent that inhibits VEGF, bFGF, tumor necrosis factor-alpha (TNF-α), and various other pro-angiogenic factors.300 Thalidomide has been reported to stimulate nuclear export of β-Arrestin1 and trigger aberrant localization of HIF-1α, causing the inhibition of HIF-1-dependent transcription of VEGF-A in breast carcinoma cells.301 In addition, thalidomide has been reported to activate sphingomyelinase to generate ceramide, which plays a key role in VEGF-induced angiogenesis, resulting in repression of VEGF receptor expression.302 Thalidomide also regulates TNF-α, which could contribute to the decreased squamous cell carcinoma incidence observed in a hamster model.303 Furthermore, treatment with thalidomide caused suppression of proliferation, inflammation, and angiogenesis due to inhibition of NF-κB pathways, which were linked to angiogenesis-related cytokine transcription.304,305 However, response to thalidomide treatment for solid malignancies, including AIDS-related Kaposi’s sarcoma, gliomas, and renal carcinoma, was modest.306 In androgen-dependent prostate cancer, thalidomide showed a significant anticancer effect, with promising performance in a Phase III clinical trial.307 Cancer-related clinical trials of thalidomide have been undertaken, regardless of its toxicity issues, while awaiting the development of thalidomide-based analogs with better bioavailable and lower toxicity.
Itraconazole
Itraconazole is an FDA-approved traditional antifungal agent with an excellent safety profile.308 Unexpectedly, data from preclinical or clinical studies of itraconazole indicated potential anticancer activity in mono- or combination therapies.309–311 The anticancer mechanisms of itraconazole probably involve inhibition of resistant protein P-glycoprotein, interference with the tumor microenvironment and mediation of other tumor development-associated signaling pathways.312,313 Notably, the antiangiogenesis activity of itraconazole was identified by drug repurposing screening which accelerated its potential use for clinical cancer therapy.314 In addition, growing studies are suggesting itraconazole has a multi-target antiangiogenesis effect by regulating various angiogenic signaling pathways.315 For example, a study has suggested that itraconazole-induced mTOR inhibition could inhibit angiogenesis via the cholesterol trafficking pathway.316 Head et al. further demonstrated that itraconazole directly targeted the mitochondrial protein VDAC1 to regulate AMPK and mTOR, while itraconazole was found to bind to the sterol-sensing domain of NPC1, a lysosomal protein closely associating with cholesterol trafficking, resulting in comprehensive inhibition of cell proliferation and angiogenesis.317 Recently, Chen et al. showed that itraconazole significantly inhibited angiogenesis in infantile hemangioma by downregulation of the PDGF/PI3K/Akt/mTOR pathway.318 Such studies have led to increased itraconazole-based clinical usage in the past few years. Studies have implied that itraconazole treatment has benefited many patients with ovarian, pancreatic and other cancers.319–322 Interestingly, a Phase II clinical study demonstrated that itraconazole combined with standard chemotherapy (pemetrexed) significantly promoted both progression free and overall survival of lung cancer patients, suggesting that the antiangiogenic properties contribute to satisfactory outcomes.309 However, some studies have implied certain contraindications for itraconazole, especially interference with other cancer drugs, like rituximab.323,324 Despite this, itraconazole is still a promising anticancer agent for clinical application because of existing preclinical or clinical data and drug safety profiles.
Non-oncology drugs which work by suppression of invasion and metastasis
Tumor invasion is the mechanism by which tumor cells spread to the surrounding environment, while tumor metastasis is where cancer cells leave the primary tumor and migrate to a new location where they generate new (secondary) tumors. These activities involve regulating existing cellular mechanisms, such as adherens junction signal transduction pathways.325 In contrast, defects in these processes can systemically cause tissue abnormalities and disrupt homeostasis, leading to genetic abnormalities or invasion and metastasis of tumors. Accordingly, more and more existing drugs with potent activity against metastatic cancer are being identified by drug repurposing, with the potential of greatly improving the survival of critically ill patients.
Berberine
Berberine is an isoquinoline alkaloid present in plants. It is a traditional Chinese medicine and was originally used for the treatment of bacterial diarrhea.326 With an increased understanding of berberine, its effect on down-regulation of lipid levels was found to relate to the inhibition of HMG-CoA reductase with benefits for patients with nonalcoholic fatty liver.327,328 In addition, berberine was reported to reduce glycemic index by enhancing insulin action or restoring insulin sensitivity, by inhibiting AMPK activation, or upregulating InsR expression, respectively.329–331 Recently, numerous studies have indicated that berberine exerts anticancer activity by preventing invasion and metastasis in breast and colorectal cancer and other malignancies. For example, a previous study indicated that berberine modulated ephrin-B2 and inhibited MMP-2 and MMP-9 expression by downregulating TGF-β1, resulting in repression of cell proliferation and metastasis in breast cancer.332 A similar outcome from a parallel study showed that berberine inhibited the expression of MMP2 and MMP9 by the regulation of the COX-2/PGE2–JAK2/STAT3 axis.333 In addition, berberine inhibited EMT by the inhibition of the RARα/β-mediated PI3K/AKT signaling pathway in melanoma.334 In endometrial cancer, berberine was shown to inhibit migration, invasion and metastasis by transcriptionally upregulating miR-101, resulting in inhibition of COX-2/prostaglandin E2 (PGE2) signaling pathways.335 Interestingly, berberine can also mediate angiogenesis, whose activation is closely linked to cancer metastasis, by directly reducing VEGF mRNA expression, inhibiting HIF-1α-induced VEGF expression or compromising the PI3K/AKT signaling pathway.336,337 Berberine has been used in clinic as an antidiabetic or hypolipidemic agent that has attracted substantial attention.338–340 Regardless of poor bioavailable by oral administration, drug repurposing of berberine as a potential anticancer agent is being currently investigated in clinical trials (NCT03281096) (NCT03333265).
Niclosamide
Niclosamide is an FDA-approved antihelminthic drug that has been listed as one of the most crucial drugs by the World Health Organization (WHO).341 Mechanistically, niclosamide treatment can inhibit glucose uptake and anaerobic metabolism of cells, suggesting a potential effect against cancer cells by targeting tumor metabolism.342 Recently, niclosamide has been shown in a number of studies to exert anticancer activity in colorectal, breast and ovarian cancer, and other malignancies.343–346 The mechanisms of the niclosamide-induced anticancer effect may be related to multiple signaling pathways, such as Wnt/β-catenin, STAT3, and NF-κB, contributing to cancer invasion and metastasis.347,348 In colorectal cancer, a calcium-binding protein S100A4 was found to be upregulated and to promote cancer metastasis.344,349 Niclosamide can inhibit liver metastasis of colorectal cancer cells by downregulation of S100A4, causing abrogation of the Wnt/β-catenin signaling pathway.344,349 Consistent with this, niclosamide has been shown to reduce lung cancer invasion by inhibition of the S100A4/NF-κB/MMP9 axis.350 A similar study reported that niclosamide impaired pulmonary metastases of breast cancer cells by reduction of the STAT3-FAK-Src axis.351 In addition, niclosamide can reduce the metastatic potential of lapatinib-resistance breast cancer cell by inhibiting EMT and alleviating stemness phenotype.352 Niclosamide in enzalutamide-resistant advanced prostate cancer cells reversed drug resistance, migration and invasion by inhibiting IL6-STAT3-AR signaling pathways.353 Data from prostate cancer cells following niclosamide treatment indicated that it inhibited tumor invasion by promoting juxtanuclear lysosome aggregation mediated by lysosome acidification.354 An increasing number of studies have shown niclosamide has an anti-metastasis effect in oral squamous cell carcinoma, hepatocellular carcinoma, melanoma, and human thyroid cancer.355–358 A Phase II clinical trial of niclosamide, in which the drug was applied to patients with colorectal cancer metastases, has been conducted to assess the drug safety and efficacy (NCT02519582). Clinical trials have also been conducted for the treatment of prostate cancer (NCT02532114) (NCT03123978). However, the poor water solubility and bioavailability of niclosamide has hindered its further clinical development for cancer therapy.359 Systemic intravenous administration of niclosamide may improve future applications in the clinic.360,361
Non-oncology drugs which work by disruption of DNA damage response
Genome instability is one of the major characteristics of malignancy, allowing certain favorable mutant tumor genotypes to survive under stress conditions.37,362 Recently, epigenetic regulation, such as DNA, RNA, and histone modification, have also been shown to enable tumors to acquire growth advantages leading to genome instability.363–365 Traditional cancer therapies are still focused on chemotherapy and radiotherapy, which induce genotoxic stress by triggering DNA damage. However, these therapies result in severe side effects and can stimulate tumors to develop drug resistance or cancer metastasis even if there is brief tumor dissipation. These worse outcomes result from genome instability, inducing abnormal activation of caretaker genes required for DNA maintenance, which are involved in detection and reparation of DNA damage.366–368 Accordingly, a method for enhancing the therapeutic index of traditional cancer therapies could be repurposing of drugs as sensitizers of genotoxic therapy to directly inhibit DNA damage response.
Spironolactone
Spironolactone, an aldosterone inhibitor originally used as an effective diuretic for the treatment of high blood pressure and edema, was subsequently shown to benefit patients with heart failure.369–371 Increasingly, studies have shown the anticancer properties of spironolactone on prostate and breast cancer.372,373 Regulation of DNA damage response appears to be the key mechanism of spironolactone in cancer treatment. For example, spironolactone was identified as an effective nucleotide excision repair inhibitor by drug repurposing, promoting chemosensitivity in cancer cells treated with platinum-based drugs. This is due to spironolactone reversibly inducing the degradation of XPB in a ubiquitin-activating enzyme- and proteasome-dependent manner, impairing both basal and activated mRNA transcription of the transcription/repair factor TFIIH.374,375 In agreement with this, a recent study also demonstrated that spironolactone could inhibit SIRT2-mediated transcription-coupled nucleotide excision repair, disturbing cisplatin-induced DNA crosslinks in lung cancer.376 In addition, the function of spironolactone on reducing homology directed repair frequencies was confirmed. Goldberg et al. found that spironolactone promoted activity of various drugs targeting DNA damage, such as Poly (ADP-ribose) polymerase (PARP) inhibitor and chemotherapeutic drugs involved in DNA cross-linking, by the impairment of Rad51 foci formation.377 They also identified that spironolactone could influence DNA double-strand break repair in cancer stem cells.378 Interestingly, Guillotin et al. showed another diuretic drug, triamterene, was active against DNA mismatch repair-deficient tumors. They reported that triamterene increased reactive oxygen species (ROS)-mediated DNA double strand breaks by regulating folate synthesis enzyme and thymidylate synthase.379 To date, clinical trials of both spironolactone and triamterene, based on mediating DNA damage response, are rare. However, population-wide data from the Prostate Cancer Data Base Sweden indicated a close relationship between spironolactone treatment and reduced prostate cancer risk.373 Taken together, spironolactone and triamterene identified by drug repurposing have shown favorable potential for clinical trials using monotherapy or combinational treatment, mediating DNA damage response for cancer therapy.
Mebendazole
Mebendazole, a synthetic benzimidazole, was approved by the FDA for treating intestinal helminthiasis by targeting tubulin polymerization.380 In the early 2000s, mebendazole was report to act against non-small cell lung cancers by tubulin depolymerization-induced cell cycle arrest.381 Subsequently, tubulin polymerization, the original target of mebendazole, was proposed as its main mechanism of action against glioblastoma and gastric cancer.382,383 Importantly, it was shown that mebendazole sensitized cancer cells to radiotherapy by regulating DNA damage response proteins. Markowitz et al. reported that mebendazole inhibited the translocation of Chk2 and Nbs1, mediators of DNA double-strand break repair, to the nucleus, resulting in an increasing effect of radiotherapy.384 In addition, mebendazole was shown to have radio-sensitizing activity in breast cancer, and decreased the fraction of stem-like cells via the hedgehog pathway, probably regulating glioma-associated oncogene homolog 1 (GLI1) (the downstream effector of the hedgehog pathway) to respond to DNA damage.385–387 In further agreement, a recent study reported that mebendazole reduced GLI1 expression in advanced thyroid cancer.388 Mebendazole was also shown to have anticancer activity and enhanced cisplatin-induced DNA damage, by inhibition of DNA double-strand break repair, in head and neck squamous cell carcinoma.389 Interestingly, benzimidazole derivatives were reported as potential dual inhibitors for PARP-1 and DHODH, two key proteins involved in DNA replication and repair mechanisms, respectively.390 To date, several clinical trials investigating the use of mebendazole for cancer therapy have addressed brain tumors, such as high-grade glioma, medulloblastoma, and astrocytoma (NCT02644291) (NCT01729260), which were mainly treated by radiotherapy or chemotherapy. In addition, a clinical trial using combinational treatment with chemotherapeutic agents (vincristine and carboplatin) and mebendazole was conducted for treating pediatric gliomas in Cohen Children's Medical Center of New York (NCT01837862). A similar clinical trial was run for patients with colorectal cancer (NCT03925662). These studies confirmed the potential effect of mebendazole on inhibiting DNA damage repair-related mechanisms. Two case reports of mebendazole treatment in adrenocortical carcinoma and colon cancer have now been published.391,392 These data showed marked remission of cancer metastasis with mebendazole treatment with no obvious adverse effects. The safety, low price, and proven anticancer effect of mebendazole support the feasibility of its clinical application as an adjuvant therapy in multiple cancers.
Non-oncology drugs which work by targeting tumor-promoting inflammation
Inflammation is typically associated with the whole process of tumorigenesis, supporting and accelerating the progression of the incipient neoplasm into a malignant tumor by generating various bioactive molecules that can influence the microenvironment around the cancer cells.393 In general, growth or survival factors, which maintain an active state of tumor growth by inducing proliferative signaling or reducing cell death are released.394,395 In addition, both angiogenic factors and metastasis-related proteins support angiogenesis, invasion and metastasis, resulting from the induction of inflammation.37,395 Inflammation can also induce the generation of chemicals, like ROS, which may act as a second messenger to mediate certain signaling pathways in cancer cells, accelerating their genetic evolution, and facilitating their acquisition of the basic hallmarks of cancer.396 Regulation of inflammation is therefore an actionable strategy for delaying tumor development and improving cancer therapy efficacy. Many existing drugs identified by drug repurposing exert their anticancer properties by mediating inflammation processes.
Aspirin
Acetylsalicylic acid, commonly known as aspirin, is a non-steroidal anti-inflammatory drug (NSAID) approved by the FDA for the treatment of pain and fever and subsequently in the treatment of stroke and cardiovascular disease.397–399 Aspirin has also attracted attention for its potential use in cancer therapy. The first suggestion that aspirin may have potential anticancer effects was reported almost 50 years ago, as evidenced by the reduction of lung metastases in aspirin-treated tumor-bearing mice.400 Multiple studies have investigated the anticancer effect of aspirin in various cancers, and explained the properties by multiple molecular mechanisms. For example, studies have indicated that aspirin has inhibitory effects on COX-1/2, which are closely associated with cancer-related inflammation by stimulating the synthesis of various prostaglandins, such as PGE2.401,402 Subsequently, aspirin was demonstrated to kill melanoma, prostate, ovarian and other cancers by inhibition of PGE2 expression.403–406 Aspirin was also reported to mediate tumor-promoting inflammation in a COX independent manner, mainly by regulating the NF-κB signaling pathway, which was initially shown as a key event relating to chronic inflammation and increased cancer risk.407 Zhang et al. showed that some aspirin-based drugs exerted anticancer activity in colon and pancreatic cancer cells by inducing ANXA1, which could prevent NF-κB binding to DNA leading to increased apoptosis and decreased proliferation.408 Furthermore, Huo et al. recently found that aspirin prevented NF-κB activation and CDX2 expression stimulated by acid and bile salts in esophageal squamous cells from patients with Barrett's esophagus.409 Aspirin treatment was also reported to enhance COMMD1 acetylation, regulating the ubiquitylation and nucleolar translocation of the RelA NF-κB subunit.410 Aspirin can also mediate the production of inflammatory cytokines, such as MCP-1 adipokine and interleukin (IL)-6.411,412 Interestingly, aspirin-triggered resolvins and lipoxins, as the specialized pro-resolving mediators, induce tumor cell debris degradation by macrophage phagocytosis and antagonize macrophage-secreted pro-inflammatory cytokines.413 Taken together, stimulating preclinical studies supports the notion that drug repurposing of aspirin as an anticancer agent for cancer prevention or treatment has potential. However, aspirin intake must be carefully controlled as it can cause intracerebral and gastrointestinal bleeding, a barrier to the widespread use of aspirin.414 Recently, Stegeman et al. demonstrated that people between age 40 and 85 would benefit from aspirin treatment designed to prevent a primary cancer.415 In addition, a previous study indicated that a protective effect against gastrointestinal, esophageal, pancreatic, brain, and lung cancer was associated with a low dose of aspirin (75 mg/day).416 Subsequently, several studies suggested a favorable effect of aspirin on colorectal cancer.417,418 Notably, Li et al. indicated patients with colorectal cancer, especially with positive PTGS2 (COX-2) expression and PIK3CA mutation, would benefit from post-diagnosis aspirin therapy, evidenced by improved overall survival.419 Clinical trials on the use of aspirin in cancer therapy are becoming increasingly common worldwide, suggesting uptake of the routine clinical application of aspirin for cancer therapy.
Thiocolchicoside
Thiocolchicoside is a semisynthetic colchicoside derived from Gloriosa superba (Liliaceae) and is clinically approved for treating rheumatologic and orthopedic disorders due to an analgesic and anti-inflammatory effect.420 Interestingly, several studies have recently mentioned the anticancer properties of thiocolchicoside. For example, Reuter et al. reported that thiocolchicoside exerted significant anticancer activity in multiple cancers, including leukemia, myeloma, and squamous cell carcinoma. Their results indicated that thiocolchicoside reduced the activation of NF-κB and COX-2 by inducing ubiquitination degradation of IκBα, a key inhibitor of the NF-κB signaling pathway mediating IKK status and p65 nuclear translocation.421 Subsequently, they further identified the effect of thiocolchicoside on inhibition of cancer-induced bone metastasis with the ability of suppressing receptor activator of NF-κB ligand and the NF-κB signaling pathway.422 Notably, several pharmaceutical companies, provide thiocolchicoside (e.g., Muscoril and Myoril) as a myorelaxant and promote its application as an anti-inflammatory and analgesic drug.423 In addition, some clinical trials confirmed the drug safety of thiocolchicoside in the treatment of acute lower back pain.424,425 Thiocolchicoside, now a half-century old drug, therefore has substantial potential for cancer therapy by drug repurposing regardless of the few clinical trials to date investigating its anticancer activity. Interestingly, clinical trials of colchicine, which is closely related to thiocolchicoside, have been conducted in Kaohsiung Medical University Hospital to investigate its anticancer effect on invasion and metastasis in patients with hepatocellular carcinoma (NCT01935700) (NCT04264260).
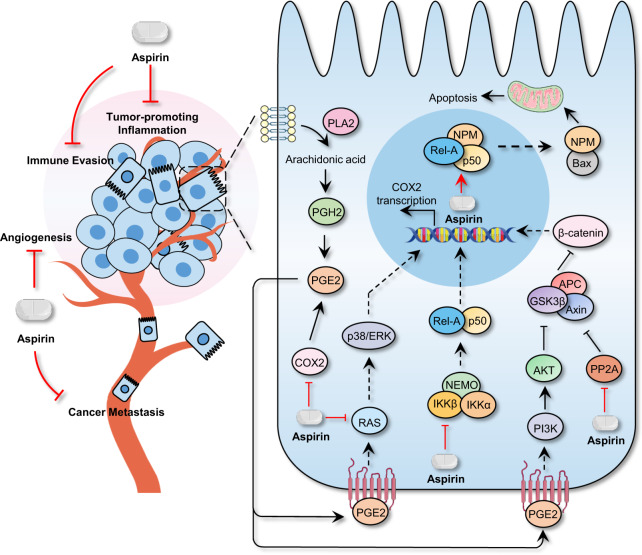
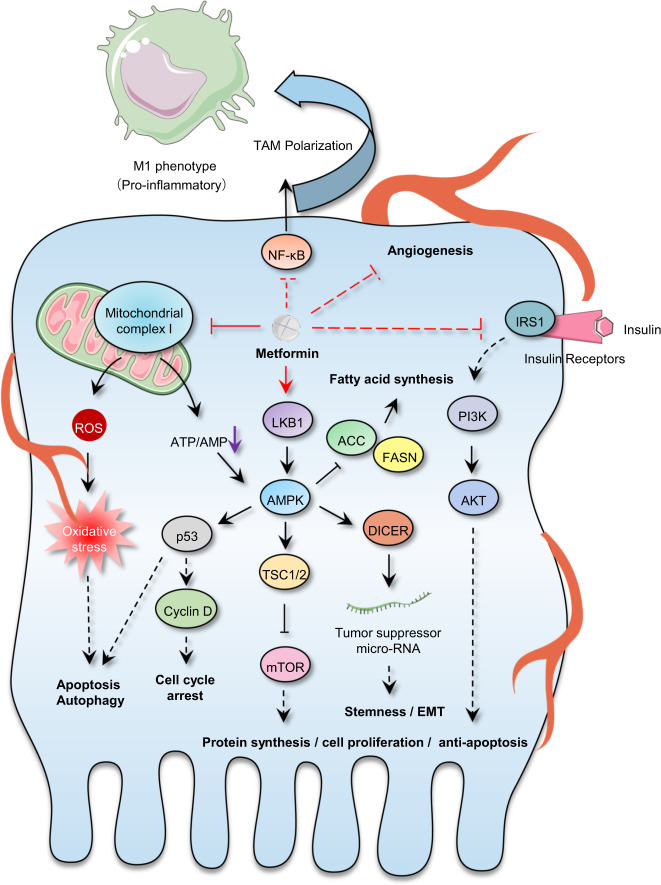
Undeniably, specially targeted drugs with powerful anticancer activity and relatively few off-target effects are responsible for improved patient survival in the clinic.14 However, multiple lines of evidence suggest that most of these molecular agents show a transitory tumor therapeutic effect ultimately leading to relapse, because several alternative parallel signaling pathways supporting malignancy can be activated attributing to adaptation of therapy pressure.37,238,426,427 In this section, we summarize the potential existing drugs, that were investigated for cancer therapy in ongoing or forthcoming clinical trials, based on the hallmarks of cancer.37,238,426,427 Given the shortcoming of targeted drugs that stimulate adaptive resistance, discovery of traditional non-oncology compounds that exert comprehensive inhibitory effect on more than one characteristic hallmark is ideal for developing durable strategies for cancer therapy by drug repurposing. This strategy has already shown efficacy. For instance, aspirin can directly regulate different target proteins, such as COXs, PP2A, RAS, IKKβ, and histone, acting on multiple hallmarks of cancer including tumor-promoting inflammation, deregulating energy metabolism, angiogenesis, cancer metastasis, and immune evasion (Fig. 3).428 Metformin has been identified to have potential for mediating various pathways, including insulin/IGF1, NF-κB, AMPK/mTOR/PI3K, Ras/Raf/Erk, Wnt, Notch, and TGF-β signaling, therefore regulating cell proliferation, self-renewal, cancer metastasis, angiogenesis, and energy metabolism (Fig. 4).429 Similar outcomes have been obtained with disulfiram, itraconazole, and artemisinin.54,312,428,430
Fig. 3.
Signaling pathways mediated by aspirin. Multiple signaling pathways regulated by aspirin is shown, acting on diverse hallmarks of cancer including tumor-promoting inflammation, deregulating energy metabolism, angiogenesis, cancer metastasis and immune evasion, are shown
Fig. 4.
Signaling pathways mediated by metformin. Direct or indirect protein targeting by Metformin is shown. These influences diverse hallmarks of cancer including regulating cell proliferation, self-renewal, cancer metastasis, angiogenesis and energy metabolism
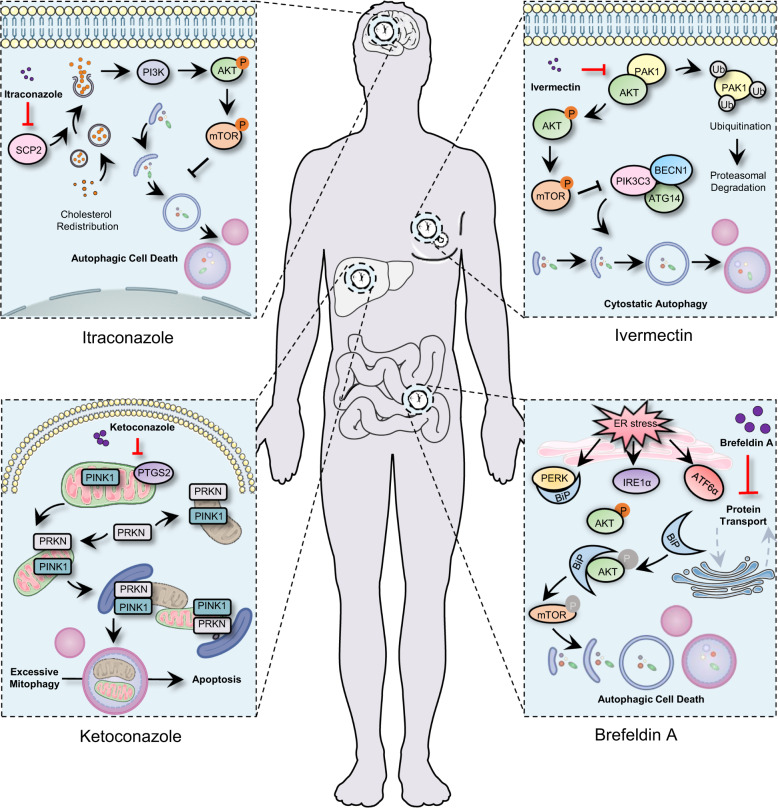
Indeed, previous studies in our group have repurposed a panel of non-oncology drugs for cancer treatment, including antifungal agents (ciclopirox olamine, ketoconazole and itraconazole), antiparasitic drug (ivermectin), anthelminthic drug (pyrvinium), antibiotic agent (brefeldin A), and natural products (quercetin, 3′-epi-12β-hydroxyfroside and toxicarioside O), showing potential promises for drug repurposing in future cancer therapy (Fig. 5). For example, brefeldin A, a macrolide antibiotic, showed significant autophagic cell death both in vitro and in vivo in colorectal cancer. Mechanistically, brefeldin A provoked endoplasmic reticulum stress-mediated upregulation of binding immunoglobulin protein (Bip), therefore promoting to increased Bip/Akt interaction resulting in decreased Akt phosphorylation, which usually activates autophagy.431 Another study found that ketoconazole, a traditional antifungal drug, induced PINK1/Parkin-mediated mitophagy by downregulating COX-2, which resulted in the acceleration of apoptosis, thereby inhibiting the growth of HCC.432 Ivermectin, a broad-spectrum antiparasitic drug swept the 2015 Nobel prize for physiology or medicine, displayed anticancer activity against breast cancer cells by inducing PAK1/Akt regulated cytostatic autophagy.433 Interestingly, recent studies demonstrate that ivermectin also inhibits the replication of SARS-CoV-2 in vitro, although the paper by Caly et al.434,435 has elicited two letters to the editor to question the clinic use for human therapy. In summary, our previous findings, together with others, have laid a solid basis for repurposed non-oncology drugs for cancer treatment. We believe this strategy will achieve great success in cancer drug development.
Fig. 5.
Partially repurposed drugs emanating from our laboratory. In addition to itraconazole, ivermectin, ketoconazole and brefeldin A which target brain, breast, liver and colon cancer, respectively, antifungal agent ciclopirox olamine, anthelminthic drug pyrvinium, and natural products quercetin, 3′-epi-12β-hydroxyfroside and toxicarioside O have also been studied
Technological approaches to drug repurposing for cancer therapy
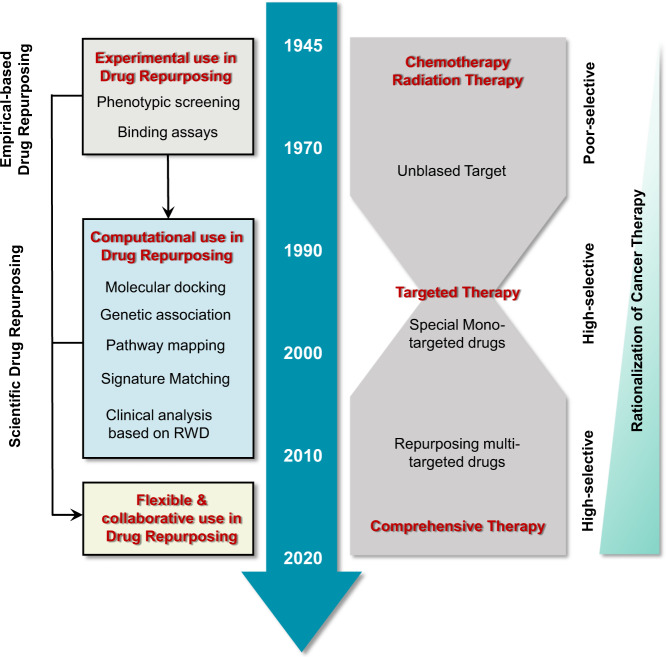
Before an old drug can be selected for evaluation as an anticarcinogen with effectiveness and therapeutic potential, a mechanistic assessment of the drug effect in preclinical models is critical and requires systematic approaches.109,436 These approaches can be broadly divided into computational and experimental approaches, which employ existing data or biochemical experiments respectively to analyze the possibility that the “old drugs can treat tumors with new tricks”.437,438 In fact, success in drug repurposing typically depends on a flexible and collaborative use of both approaches (Fig. 6).439,440 The most commonly used computational and experimental approaches and examples of drug repurposing cases are detailed below.
Fig. 6.
The evolution of screening and therapeutic strategies in drug repurposing for cancer. With the development of biochemical techniques and bioinformatics, treatments based on targeted therapy mainly focus on computational approaches of drug repurposing. Currently, drug repurposing typically depends on a flexible and collaborative use of both experimental and computational approaches to screen multi-targeted agents
Computational approaches based on molecular theory (MOT)
Understanding a drug at the molecular level, and matching it with clinical symptoms of neoplasms different from those for which it was originally approved or developed, is key for computational approaches based on MOT leading to drug repurposing.441–443 Such approaches involve comprehensive analysis of experimental data, such as chemical or protein structure, gene or protein expression, and various omics data, which assist researchers to put forward a repurposing hypothesis.444–447
Molecular structure analysis
Using high-throughput computational structural analysis is an efficient way to predict binding site complementarity between the ligand and the target.439,448 Docking studies are the main algorithmic approach for predicting the orientation of a ligand into a cavity of the target protein.449,450 High-quality structural data of known receptor targets involved in cancer can accurately predict specific drug targets using drug libraries.451,452 As an exemplar, bromovinyldeoxyuridine (BVDU) is a thymidine analog used for the treatment of herpes zoster infection, which interacts with viral thymidine kinase using several specific noncovalent binding sites.453,454 Heinrich et al. found that heat shock protein HSP27 was a potential target of BVDU, based on a predicted binding site to a viral thymidine kinase and identified some kinase inhibitors targeting HSP27 by in silico docking of a targeted library.455,456 Following up on these findings, Salentin et al. used the Protein Data Bank, which contains structural data for more than one thousand different drug targets, to explore any drug–target pairs, which have similar interactions with BVDU and thymidine kinase.457,458 The antimalarial drug amodiaquine was found to inhibit HSP27 chaperone function and reverse tumor drug resistance in multiple myeloma cell lines. Binding site comparisons are also a popular approach to predict similar pharmacological drugs.453 Lim et al. predicted the heart failure drug levosimendan as an inhibitor of RIOK1 and other kinases by using ligand binding site comparison and protein-ligand docking. They went on to show that levosimendan had anticancer effects by directly inhibiting RIOK1 and RNA processing enzymes.459
Signature or pathway matching
Various omics studies of cancer not only deepen our understanding of tumor hallmarks at the molecular level, but also provide big data for supporting drug repurposing by applying advanced bioinformatics.460,461 A detailed framework constructed by computer processing can integrate the mechanisms of drug action, phenotypes and molecular biological characteristics of cancer to identify novel drug–disease relationships and indicate potential drug repurposing possibilities.439,444 The novel associations between genes and cancer found by Genome-Wide Association Studies and Phenome-Wide Association Studies have expanded our knowledge of new targets for existing drugs.462–464 Drugs targeting cancer are inferred by overlapping cancer-associated genes and drug-targeted genes from DrugBank, a freely accessible database comprising over 13,000 drugs and related pathways or targets (https://www.drugbank.ca/).465–467 Medication Indication Resource (MEDI), another freely-available, computable MEDI can be used to confirm the plausibility of inferred drug indications, which have clinical potential.468,469 Following this approach, alosetron was discovered to have genetic correlation with bladder cancer although originally used to treat irritable bowel syndrome.470,471 Bezafibrate, a lipid regulating agent, was also suggested to have anticancer properties for melanoma.472 In a similar approach, Xu et al. utilized The Cancer Genome Atlas (TCGA) transcriptomes to construct non-tissue-specific core signatures, which were used to identify drugs whose perturbation signatures mimicked a single gene mutation.473 Combined with the use of the drug treatment profiles and an in silico screen, results indicated that several psychiatric drugs (such as trifluoperazine) and antihypertensive calcium channel blockers (such as perhexiline) might have anticancer potential by inhibition of the TGF-β pathway or modulation of AKT and AMPK phosphorylation, respectively.473,474
Computational approaches based on Real World Data (RWD)
RWD, as its name suggests, is mainly composed of the real electronic health records (EHRs) of patients characterized by large and complex datasets.475–477 EHRs are the systematized collection of electronically-stored patient and population health information in a digital format478 and contain large amounts of data on patient outcomes.479,480 These are divided into diagnostic and pathophysiological data, including experimental data, drug prescribing data, disease phenotype, and imaging data.475,481 Both structured and unstructured data from patients have considerable value as a source for drug repurposing by identifying consistent signals.482 Moreover, the plentiful amount of EHR data allows for significant statistical significance.483,484 Retrospective clinical analysis is the most commonly used computational approach based on RWD and the data are mainly obtained from EHRs.485,486 A classic case of repurposing a noncancer drug for cancer treatment from retrospective clinical analysis is that of metformin which can decrease cancer mortality in a dose dependent manner.487,488 Xu et al. designed a clinical cohort from 15 years of EHRs from Vanderbilt University Medical Center and Mayo Clinic. In the independent populations, data from over 100,000 patients with a cancer diagnosis were used in the study and were validated with beneficial effects of metformin for cancer survival, including breast, colorectal, lung, and prostate cancers.489 Valproate, previously approved for use in epilepsy, also has been successfully repurposed arising from retrospective clinical analyses for the treatment of acute myeloid leukemia and glioblastoma.490–492
Experimental approaches based on intermolecular interactions
Proteomic techniques using affinity chromatography combined with mass spectrometry, showing protein interactions based on intermolecular force have been widely used for target validation, facilitating drug repurposing due to an experimentally based pharmacological analysis.493,494 In this approach, cells or animals are treated with drugs followed by deep proteome analysis to differentially quantify proteins changes that occurred.495,496 With the development of labeling (including metabolic labeling and chemical labeling) or label-free approaches, proteins targeted by the drug can be detected in an unbiased fashion.497,498 Gefitinib, an epidermal growth factor receptor kinase inhibitor was identified to have more than 20 targets by incubating cancer cells with a covalently modified gefitinib followed by mass spectrometry.499 The Cellular Thermo Stability Assay, based on the principle of altered protein thermal stabilization/destabilization in response to ligand binding enables the identification of drug targets by estimating the interaction of target proteins and drug-like ligands.500,501 By combining this with thermal proteome profiling, which can be applied in living cells without requiring compound labeling, cellular targets can be confirmed in an unbiased manner.502–504
Approaches based on chemical genetics are also dependent on intermolecular forces, and can help explain the relationship between binding and drug efficacy.505,506 An example of the success of this technique comes from Karaman et al. They evaluated several kinase inhibitors against pathologically significant human protein kinases using a competition binding assay in vitro. Interaction maps for 38 kinase inhibitors across a panel of 317 kinases representing >50% of the predicted human protein kinome generated a heat map of 3175 binding interactions. Surprisingly, some drugs including but not limited to sorafenib showed higher affinity to other targets than the published known targets. In addition, different types of targets of existing drugs, such as the anthelmintic drug niclosamide to treat Zika virus infection, are becoming increasingly recognized due to the development of chemical genetics suggesting repurposing opportunities for cancer.507
Experimental approaches based on phenotypic screening
Phenotypic screening is a direct way of drug repurposing by analyzing the relative effects in a designed model even if previous studies have not identified the candidate drugs targets.8,44 Typically, a series of cell based in vitro assays in 96- or 384-well format are used in phenotypic screening.508 For example, Iljin et al. identified disulfiram used for alcohol abuse as an antineoplastic agent by conducting high-throughput cell-based screening, using proliferation as the phenotypic criteria, with a library of 4910 druggable small molecules against four prostate cancer and two nonmalignant prostate epithelial cell lines. Subsequently, they validated the anticancer effect of disulfiram using genome wide-gene expression studies.509 Recently, Corsello et al. reported their studies on drug repurposing from a viability evaluation of 578 human cancer cell lines of diverse tumor types treated with more than 4500 drugs including hundreds of non-oncology compounds. They utilized a novel method named “PRISM” (profiling relative inhibition simultaneously in mixtures), which tracks the proliferation of mixtures of cancer cell lines during treatment with the candidate drug using unique molecular barcodes. If a particular drug has efficacy, proliferation of cell lines will decrease, resulting in depletion of a specific molecular barcodes. Using a bead-based assay, the relative abundance of each barcode was determined and anticancer activity profiles generated. 49 non-oncology drugs had an unexpectedly high rate of anticancer activity.510,511 Whole animal screening assays can also be utilized in drug repurposing. Whole organism phenotypic assays not only identified candidate drugs effective against cancer, but additionally yielded pharmacokinetic and organ-toxicity results compared with cell-based screening.512–515 Ridges et al. used genetically engineered T-cell reporting zebrafish as a platform to evaluate over 26,000 small molecules for efficacy against leukemia and found that 1H-indole-3-carbaldehyde 8-quinolinylhydrazone, termed lenaldekar, showed significant activity against various hematologic malignancies, including T-acute lymphoblastic leukemia and chronic myelogenous leukemia.513,514
Challenges in the development of drug repurposing
Drug repurposing, aiming to repurpose old compounds for new indications, is already in widespread use in cancer therapy. However, few repurposed drugs are officially included in the published cancer clinical practice guidelines.16,516,517 In spite of advantages such as demonstrated anticancer pharmacokinetic properties, and acceptable safety and tolerability in humans, there is (as in all drug development) still a possibility of failure in later stages of clinical trials because of competition from successful new drug development.16,517 Other barriers include legal and regulatory issues such as patent-related considerations and inequitable prescription charges.16,516,517 Hopefully such barriers will prove surmountable.
Patent considerations
A repurposed product for cancer therapy could bring substantial profit to the patentee. However, the intellectual property status of these candidates is usually ambiguous and unpatentable as the scientific literature or clinical practice is prior art. Accordingly, only non-profit organizations are interested in developing such drugs as returns on investment could be low. In the Repurposing Drugs in Oncology (ReDo) project that includes 72 drugs involved in 190 registered clinical trials, <5% are sponsored by the pharmaceutical companies with the non-profit organizations represented by universities or hospitals supporting the remainder.517,518 For drugs that are off-patent, a new method-of-use patent can be obtained for a new repurposed use of an old generic drug. However, the patent application process is strict, requiring detailed data showing proof of credible treatment for the concerned indication, probably using unique formulations and dosage forms. Furthermore, if the generic manufacturer challenges those patents lengthy and costly delays may ensue. Consequently, patent consideration-induced market exclusivity has been identified as a key hurdle in drug repurposing.16 Recently, the Off‑Patent Drugs Bill 2015–16, was put before the UK Parliament to address this. It was supported by a number of medical charities but failed to pass into legislation.16
Inequitable prescription
Theoretically, drugs prescribed by physicians should take into account scientific evidence based on clinical trials, generic or repurposed drugs should be prescribed where appropriate. However, the pharmaceutical industry can influence this by spending large amounts of money on drug promotion, marketing to physicians and advertising to consumers. Thalidomide, discussed above, is an excellent example of problems that can occur. Originally prescribed as a sedative or an antiemetic with low-cost and safe properties, it was recently repurposed to treat multiple myeloma. In two randomized clinical trials, the therapeutic regimen melphalan–prednisone–lenalidomide was compared with that of melphalan–prednisone–thalidomide, and showed no survival advantage.519,520 However, lenalidomide rather than thalidomide was approved as the standard therapy even if its approximate cost is 43-fold times higher than thalidomide. Obviously, prescribing cheaper drugs, with equivalent activity to the expensive ones, benefit patients rather than biopharma.
Conclusions and future pespectives
Cancer is still the second leading cause of death globally (around 9.6 million deaths in 2018) in spite of extensive studies to find new treatment regimens and more effective drugs.1 However, the cost of cancer treatment is increasing, largely due to the expense of taking drugs through clinical trials (typically around $2.6 billion according to a recent study) where there is only a low rate of success (around 12%).521 There therefore exists an urgent need to develop effective, safe, cheaper, and readily available anticancer agents. Theoretically, drug repurposing could break the current drug shortage bottleneck. Importantly, our understanding of cancer biology and the associated hallmarks of cancer is increasing.37,238,426,427 This, coupled with repurposing studies that apply systematic screening of the entire pharmacopoeia coupled with advanced bioinformatics, should identify new drugs and their targets. In addition, the use of non-oncology drugs, that account for the bulk of our medications, has the potential to further accelerate the progress of drug repurposing. In this review we have overviewed the most commonly used approaches for drug repurposing, addressing both phenotypic-based and target-based strategies. Based on widespread recognition of these concepts and related studies of drug repurposing over the last decade, we have summarized and evaluated old non-oncology drugs as potential candidates for drug repurposing (Table 1). We have discussed how some of these drugs effectively regulate at least one hallmark of cancer, often induced by multiple parallel signaling pathways, while others exert comprehensive anticancer effects by regulating multiple targets mediated by various alternative signaling routes. Their potential for mono- or drug combination therapy is discussed with respect to the key hallmarks of cancer.
Table 1.
A total panel of repurposed drugs mentioned in this review
| Cancer hallmarks | Repurposed drug | Original application | Reported targets/pathways | Clinical trials of cancer |
|---|---|---|---|---|
| Sustaining proliferative signaling | Rapamycin | Immunosuppressant, anti-restenosis agent | mTOR and associated signaling networks | Rectum, breast, prostate cancer etc. |
| Sustaining proliferative signaling | Prazosin | Hypertension | PKCδ-dependent AKT signaling pathway | Adrenal incidentalomas |
| Sustaining proliferative signaling | Indomethacin | Rheumatic disease | Shc-ERK axis, PKCζ-p38-DRP1 axis, Wnt/β-catenin | Colorectal, esophageal, ovarian cancer etc. |
| Evading growth suppressors | Quinacrine | Malaria, giardiasis, rheumatoid arthritis | p53, FACT-CK2-p53 axis | Prostatic, non-small cell lung cancer etc. |
| Evading growth suppressors | Ritonavir | Human immunodeficiency virus | p53, CDKs-RB axis, AKT-E2F-1-RB axis | Breast cancer, Kaposi’s sarcoma etc. |
| Resisting cell death | Artemisinin and related-derivatives | Malaria | Ferroptosis, autophagy, oncosis, anoikis | Breast, colorectal, lung cancer etc. |
| Resisting cell death | Chloroquine and related-derivatives | Malaria, rheumatoid arthritis | Autophagy, PPT1 | Pancreatic, breast cancer, chondrosarcoma etc. |
| Enabling replicative immortality | Curcumin | Dermatological diseases | hTERT, Wnt/β-catenin, Hippo/YAP | Breast, prostate cancer, multiple myeloma etc. |
| Enabling replicative immortality | Genistein | Menopause, osteoporosis, obesity | hTERT, Wnt/β-catenin | Colorectal, bladder, breast cancer etc. |
| Inducing angiogenesis | Thalidomide | Sedative, antiemetic | Various proangiogenic factors, VEGF receptor, NF-κB | Prostate, ovarian, colorectal cancer etc. |
| Inducing angiogenesis | Itraconazole | Antifungal agent | mTOR-cholesterol trafficking, VDAC1, PDGF-Akt–mTOR axis | Prostate, lung cancer etc. |
| Activating invasion and metastasis | Berberine | Bacterial diarrhea | Ephrin-B2, MMP-2/MMP-9, EMT, miR-101, VEGF | Gastric, colorectal, lung cancer etc. |
| Activating invasion and metastasis | Niclosamide | Antihelminthic drug | Wnt/β-catenin, STAT3, NF-κB | Colorectal, prostate cancer etc. |
| Genome instability and mutation | Triamterene | Diuretic | Nucleotide excision repair, thymidylate synthase | Acute myelocytic leukemia etc. |
| Genome instability and mutation | Mebendazole and related-derivatives | Intestinal helminthiasis | Chk2, Nbs1, PARP-1, DHODH | Medulloblastoma, glioma, astrocytoma etc. |
| Tumor-promoting inflammation | Aspirin | Pain, fever | COX-1/2, ANXA1-NF–κB axis, CDX2, COMMD1–RelA axis | Gastrointestinal, esophageal cancer etc. |
| Tumor-promoting inflammation | Thiocolchicoside | Rheumatologic, orthopedic disorders | NF-κB-related pathways, COX-2 | None |
| Reprogramming energy metabolism | Metformin | Obese type 2 diabetes | AMPK, PI3K-mTOR pathways, BACH1 | Prostate, breast, colorectal cancer etc. |
| Reprogramming energy metabolism | Disulfiram | Alcohol-aversion drug | ALDH, NAD+-dependent proteins | Prostate, breast cancer, melanoma etc. |
| Evading immune destruction | Rotavirus vaccines, Live 17D | Infectious disease | NF-κΒ, Type I interferon pathways, CTLs, Tregs | None |
This table lists existing non-oncology agents for cancer therapy in this review, including targeting cancer hallmarks, original application, reported targets or pathways and related clinical trials in cancer treatment (https://clinicaltrials.gov/).
However, there are some key points that deserve attention. Firstly, cancer is governed by multiple, complex cellular pathways, that could ultimately inhibit the effort of a specific targeted therapy. It is not trivial to replace inoperative targeted drugs with suitable multi-targeted non-oncology drugs, as evidenced by the few regimes currently used to treat cancer patients. So where do we go next? Drug combination therapies, an alternative strategy, should be considered which might have a higher success rate in clinical application. Drug combination therapies usually target multiple mechanisms, including downstream off-target, parallel pathways or compensatory signaling that contribute to tumorigenesis. Next, current cancer treatments mainly focus on directly killing cancer cells in both de novo drug discovery and drug repurposing. This direction seems rationale because increasing programmed cell death, like lethal-autophagy, ferroptosis, pyroptosis etc., are comprehensively studied and can provide alternative strategies for apoptosis-resistant cancer. Nevertheless, residual cancer cells (known as minimal residual disease), often remain after treatment, and are warning that “what does not kill me, makes me stronger”. Thus, using non-oncology drugs that have the potential to target the multiple hallmarks of cancer and the related cancer biology, instead of directly killing the cancer cells per ce heralds them as a key complement to the new paradigm of personalized/precision medicine in the coming future. Meanwhile, the ever-increasing availability of biomedical data from public databases coupled with continued advances in techniques and analytical methods forms a sound basis for the accurate identification of repurposing candidates. In this respect, the PRISM public resource repurposing dataset, an unbiased multi-platform approach recently generated by Corsello et al., will prove a starting point for many future orientation studies.510,511
In summary, drug repurposing that opened a window for drug discovery is a trend for including, but not limited to, cancer therapy, regardless of many failures. As Nobel laureate in physiology and medicine, James Black, once said: “the most fruitful basis for the discovery of a new drug is to start with an old drug”.522 We totally concur with this concept.
Acknowledgements
This work was supported by grants from the Chinese NSFC (nos. 81821002, 81790251, and 81773143), and Guangdong Basic and Applied Basic Research Foundation (2019B030302012).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Zhe Zhang, Li Zhou, Na Xie
Contributor Information
Tao Zhang, Email: ztbill@cmc.edu.cn.
Yongping Cui, Email: cuiyp@sphmc.org.
Canhua Huang, Email: hcanhua@scu.edu.cn.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, et al. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Kirsch J, et al. Biosensor technology: recent advances in threat agent detection and medicine. Chem. Soc. Rev. 2013;42:8733–8768. doi: 10.1039/c3cs60141b. [DOI] [PubMed] [Google Scholar]
- 4.Shaked Y. The pro-tumorigenic host response to cancer therapies. Nat. Rev. Cancer. 2019;19:667–685. doi: 10.1038/s41568-019-0209-6. [DOI] [PubMed] [Google Scholar]
- 5.Eder J, Sedrani R, Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 2014;13:577–587. doi: 10.1038/nrd4336. [DOI] [PubMed] [Google Scholar]
- 6.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 7.Mullard A. Partnering between pharma peers on the rise. Nat. Rev. Drug Discov. 2011;10:561–562. doi: 10.1038/nrd3526. [DOI] [PubMed] [Google Scholar]
- 8.Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery—past, present and future. Nat. Rev. Drug Discov. 2014;13:588–602. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 9.Su M, et al. Availability, cost, and prescription patterns of antihypertensive medications in primary health care in China: a nationwide cross-sectional survey. Lancet. 2017;390:2559–2568. doi: 10.1016/S0140-6736(17)32476-5. [DOI] [PubMed] [Google Scholar]
- 10.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 11.Kurzrock R, Kantarjian HM, Kesselheim AS, Sigal EV. New drug approvals in oncology. Nat. Rev. Clin. Oncol. 2020;17:140–146. doi: 10.1038/s41571-019-0313-2. [DOI] [PubMed] [Google Scholar]
- 12.Paul SM, et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 13.Petsko GA. When failure should be the option. BMC Biol. 2010;8:61. doi: 10.1186/1741-7007-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedard PL, Hyman DM, Davids MS, Siu LL. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet. 2020;395:1078–1088. doi: 10.1016/S0140-6736(20)30164-1. [DOI] [PubMed] [Google Scholar]
- 15.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushpakom S, et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 17.Pantziarka P. Scientific advice—is drug repurposing missing a trick? Nat. Rev. Clin. Oncol. 2017;14:455–456. doi: 10.1038/nrclinonc.2017.69. [DOI] [PubMed] [Google Scholar]
- 18.Corsello SM, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clohessy JG, Pandolfi PP. Mouse hospital and co-clinical trial project-from bench to bedside. Nat. Rev. Clin. Oncol. 2015;12:491–498. doi: 10.1038/nrclinonc.2015.62. [DOI] [PubMed] [Google Scholar]
- 20.Maxmen A. Busting the billion-dollar myth: how to slash the cost of drug development. Nature. 2016;536:388–390. doi: 10.1038/536388a. [DOI] [PubMed] [Google Scholar]
- 21.Bertolini F, Sukhatme VP, Bouche G. Drug repurposing in oncology-patient and health systems opportunities. Nat. Rev. Clin. Oncol. 2015;12:732–742. doi: 10.1038/nrclinonc.2015.169. [DOI] [PubMed] [Google Scholar]
- 22.Huang A, Garraway LA, Ashworth A, Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020;19:23–38. doi: 10.1038/s41573-019-0046-z. [DOI] [PubMed] [Google Scholar]
- 23.Roumenina LT, et al. Context-dependent roles of complement in cancer. Nat. Rev. Cancer. 2019;19:698–715. doi: 10.1038/s41568-019-0210-0. [DOI] [PubMed] [Google Scholar]
- 24.Rancati G, Moffat J, Typas A, Pavelka N. Emerging and evolving concepts in gene essentiality. Nat. Rev. Genet. 2018;19:34–49. doi: 10.1038/nrg.2017.74. [DOI] [PubMed] [Google Scholar]
- 25.Tambuyzer E, et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat. Rev. Drug Discov. 2020;19:93–111. doi: 10.1038/s41573-019-0049-9. [DOI] [PubMed] [Google Scholar]
- 26.Dallavalle S, et al. Improvement of conventional anticancer drugs as new tools against multidrug resistant tumors. Drug Resist Updates. 2020;50:100682. doi: 10.1016/j.drup.2020.100682. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Dong G, Sheng C. Structural simplification of natural products. Chem. Rev. 2019;119:4180–4220. doi: 10.1021/acs.chemrev.8b00504. [DOI] [PubMed] [Google Scholar]
- 28.Patel MN, et al. Objective assessment of cancer genes for drug discovery. Nat. Rev. Drug Discov. 2013;12:35–50. doi: 10.1038/nrd3913. [DOI] [PubMed] [Google Scholar]
- 29.Swinney DC. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clin. Pharm. Ther. 2013;93:299–301. doi: 10.1038/clpt.2012.236. [DOI] [PubMed] [Google Scholar]
- 30.Moffat JG, et al. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 31.Flory J, Lipska K. Metformin in 2019. JAMA. 2019;321:1926–1927. doi: 10.1001/jama.2019.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris A. In search of the mechanisms of metformin in cancer. Nat. Rev. Endocrinol. 2018;14:628. doi: 10.1038/s41574-018-0104-3. [DOI] [PubMed] [Google Scholar]
- 33.Martin M, Marais R. Metformin: a diabetes drug for cancer, or a cancer drug for diabetics? J. Clin. Oncol. 2012;30:2698–2700. doi: 10.1200/JCO.2012.42.1677. [DOI] [PubMed] [Google Scholar]
- 34.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 35.Turner NC, Reis-Filho JS. Genetic heterogeneity and cancer drug resistance. Lancet Oncol. 2012;13:e178–e185. doi: 10.1016/S1470-2045(11)70335-7. [DOI] [PubMed] [Google Scholar]
- 36.Lee JK, et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018;50:1399–1411. doi: 10.1038/s41588-018-0209-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 38.Kelly-Irving M, Delpierre C, Vineis P. Beyond bad luck: induced mutations and hallmarks of cancer. Lancet Oncol. 2017;18:999–1000. doi: 10.1016/S1470-2045(17)30520-X. [DOI] [PubMed] [Google Scholar]
- 39.Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J. Clin. Oncol. 2013;31:1039–1049. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pauli C, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguirre AJ, et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018;8:1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubio-Perez C, et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell. 2015;27:382–396. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Cheng F, et al. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 2019;10:3476. doi: 10.1038/s41467-019-10744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M, et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019;10:3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klaeger S, et al. The target landscape of clinical kinase drugs. Science. 2017;358:eaan4368. doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katsnelson A. Drug development: target practice. Nature. 2013;498:S8–S9. doi: 10.1038/498S8a. [DOI] [PubMed] [Google Scholar]
- 49.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:eaal2380. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarmento-Ribeiro AB, et al. The emergence of drug resistance to targeted cancer therapies: clinical evidence. Drug Resist Updat. 2019;47:100646. doi: 10.1016/j.drup.2019.100646. [DOI] [PubMed] [Google Scholar]
- 51.Boyer A, et al. Drug repurposing in malignant pleural mesothelioma: a breath of fresh air? Eur. Respir. Rev. 2018;27:170098. doi: 10.1183/16000617.0098-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang QI, et al. Preclinical pharmacodynamic evaluation of antibiotic nitroxoline for anticancer drug repurposing. Oncol. Lett. 2016;11:3265–3272. doi: 10.3892/ol.2016.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hernandez JJ, et al. Giving drugs a second chance: overcoming regulatory and financial hurdles in repurposing approved drugs as cancer therapeutics. Front Oncol. 2017;7:273. doi: 10.3389/fonc.2017.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Efferth T. From ancient herb to modern drug: artemisia annua and artemisinin for cancer therapy. Semin Cancer Biol. 2017;46:65–83. doi: 10.1016/j.semcancer.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 56.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov. 2017;7:137–155. doi: 10.1158/2159-8290.CD-16-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diamond EL, et al. Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Discov. 2016;6:154–165. doi: 10.1158/2159-8290.CD-15-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrik-Outmezguine VS, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahronian LG, et al. Clinical acquired resistance to RAF Inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 61.Sharif A. Sirolimus after kidney transplantation. BMJ. 2014;349:g6808. doi: 10.1136/bmj.g6808. [DOI] [PubMed] [Google Scholar]
- 62.Fattori R, Piva T. Drug-eluting stents in vascular intervention. Lancet. 2003;361:247–249. doi: 10.1016/S0140-6736(03)12275-1. [DOI] [PubMed] [Google Scholar]
- 63.Farb A, et al. Oral everolimus inhibits in-stent neointimal growth. Circulation. 2002;106:2379–2384. doi: 10.1161/01.cir.0000033973.06059.04. [DOI] [PubMed] [Google Scholar]
- 64.Grabiner BC, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dancey J. mTOR signaling and drug development in cancer. Nat. Rev. Clin. Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. [DOI] [PubMed] [Google Scholar]
- 66.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altman JK, et al. Dual mTORC2/mTORC1 targeting results in potent suppressive effects on acute myeloid leukemia (AML) progenitors. Clin. Cancer Res. 2011;17:4378–4388. doi: 10.1158/1078-0432.CCR-10-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown VI, et al. Rapamycin is active against B-precursor leukemia in vitro and in vivo, an effect that is modulated by IL-7-mediated signaling. Proc. Natl Acad. Sci. Usa. 2003;100:15113–15118. doi: 10.1073/pnas.2436348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sillaber C, et al. Evaluation of antileukaemic effects of rapamycin in patients with imatinib-resistant chronic myeloid leukaemia. Eur. J. Clin. Invest. 2008;38:43–52. doi: 10.1111/j.1365-2362.2007.01892.x. [DOI] [PubMed] [Google Scholar]
- 70.Mancini M, et al. RAD 001 (everolimus) prevents mTOR and Akt late re-activation in response to imatinib in chronic myeloid leukemia. J. Cell Biochem. 2010;109:320–328. doi: 10.1002/jcb.22380. [DOI] [PubMed] [Google Scholar]
- 71.O'Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choo AY, et al. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl Acad. Sci. USA. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maiso P, et al. Defining the role of TORC1/2 in multiple myeloma. Blood. 2011;118:6860–6870. doi: 10.1182/blood-2011-03-342394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McHugh DJ, et al. A phase I trial of IGF-1R inhibitor cixutumumab and mTOR inhibitor temsirolimus in metastatic castration-resistant prostate cancer. Clin. Genitourin Cancer. 2020;18:171–178.e2. doi: 10.1016/j.clgc.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rugo HS, et al. A randomized phase II trial of ridaforolimus, dalotuzumab, and exemestane compared with ridaforolimus and exemestane in patients with advanced breast cancer. Breast Cancer Res Treat. 2017;165:601–609. doi: 10.1007/s10549-017-4375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitsiades CS, Hayden PJ, Anderson KC, Richardson PG. From the bench to the bedside: emerging new treatments in multiple myeloma. Best. Pract. Res Clin. Haematol. 2007;20:797–816. doi: 10.1016/j.beha.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rao RD, et al. Disruption of parallel and converging signaling pathways contributes to the synergistic antitumor effects of simultaneous mTOR and EGFR inhibition in GBM cells. Neoplasia. 2005;7:921–929. doi: 10.1593/neo.05361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cirstea D, et al. Dual inhibition of akt/mammalian target of rapamycin pathway by nanoparticle albumin-bound-rapamycin and perifosine induces antitumor activity in multiple myeloma. Mol. Cancer Ther. 2010;9:963–975. doi: 10.1158/1535-7163.MCT-09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zibelman M, et al. Phase I study of the mTOR inhibitor ridaforolimus and the HDAC inhibitor vorinostat in advanced renal cell carcinoma and other solid tumors. Investig. N. Drugs. 2015;33:1040–1047. doi: 10.1007/s10637-015-0261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newell P, et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J. Hepatol. 2009;51:725–733. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malizzia LJ, Hsu A. Temsirolimus, an mTOR inhibitor for treatment of patients with advanced renal cell carcinoma. Clin. J. Oncol. Nurs. 2008;12:639–646. doi: 10.1188/08.CJON.639-646. [DOI] [PubMed] [Google Scholar]
- 83.Kirkendall WM, et al. Prazosin and clonidine for moderately severe hypertension. JAMA. 1978;240:2553–2556. [PubMed] [Google Scholar]
- 84.Skrott Z, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552:194–199. doi: 10.1038/nature25016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clifford GM, Farmer RD. Medical therapy for benign prostatic hyperplasia: a review of the literature. Eur. Urol. 2000;38:2–19. doi: 10.1159/000020246. [DOI] [PubMed] [Google Scholar]
- 86.Waldo R. Prazosin relieves Raynaud's vasospasm. JAMA. 1979;241:1037. [PubMed] [Google Scholar]
- 87.Lang CC, Choy AM, Rahman AR, Struthers AD. Renal effects of low dose prazosin in patients with congestive heart failure. Eur. Heart J. 1993;14:1245–1252. doi: 10.1093/eurheartj/14.9.1245. [DOI] [PubMed] [Google Scholar]
- 88.Mulvihill-Wilson J, et al. Hemodynamic and neuroendocrine responses to acute and chronic alpha-adrenergic blockade with prazosin and phenoxybenzamine. Circulation. 1983;67:383–393. doi: 10.1161/01.cir.67.2.383. [DOI] [PubMed] [Google Scholar]
- 89.Iwai-Kanai E, et al. alpha- and beta-adrenergic pathways differentially regulate cell type-specific apoptosis in rat cardiac myocytes. Circulation. 1999;100:305–311. doi: 10.1161/01.cir.100.3.305. [DOI] [PubMed] [Google Scholar]
- 90.Wallace JM, Gill DP. Prazosin in the diagnosis and treatment of pheochromocytoma. JAMA. 1978;240:2752–2753. [PubMed] [Google Scholar]
- 91.Nicholson JP, Jr, et al. Pheochromocytoma and prazosin. Ann. Intern Med. 1983;99:477–479. doi: 10.7326/0003-4819-99-4-477. [DOI] [PubMed] [Google Scholar]
- 92.Desiniotis A, Kyprianou N. Advances in the design and synthesis of prazosin derivatives over the last ten years. Exp. Opin. Ther. Targets. 2011;15:1405–1418. doi: 10.1517/14728222.2011.641534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Assad Kahn S, et al. The anti-hypertensive drug prazosin inhibits glioblastoma growth via the PKCdelta-dependent inhibition of the AKT pathway. EMBO Mol. Med. 2016;8:511–526. doi: 10.15252/emmm.201505421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Percy JS, Stephenson P, Thompson M. Indomethacin in the treatment of rheumatic diseases. Ann. Rheum. Dis. 1964;23:226–231. doi: 10.1136/ard.23.3.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guieu R, Blin O, Pouget J, Serratrice G. Analgesic effect of indomethacin shown using the nociceptive flexion reflex in humans. Ann. Rheum. Dis. 1992;51:391–393. doi: 10.1136/ard.51.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Timar J, et al. Neoadjuvant immunotherapy of oral squamous cell carcinoma modulates intratumoral CD4/CD8 ratio and tumor microenvironment: a multicenter phase II clinical trial. J. Clin. Oncol. 2005;23:3421–3432. doi: 10.1200/JCO.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 97.Janik JE, et al. Phase II trial of interleukin 1 alpha and indomethacin in treatment of metastatic melanoma. J. Natl Cancer Inst. 1996;88:44–49. doi: 10.1093/jnci/88.1.44. [DOI] [PubMed] [Google Scholar]
- 98.Chennamaneni S, Zhong B, Lama R, Su B. COX inhibitors Indomethacin and Sulindac derivatives as antiproliferative agents: synthesis, biological evaluation, and mechanism investigation. Eur. J. Med Chem. 2012;56:17–29. doi: 10.1016/j.ejmech.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 99.Touhey S, et al. Structure–activity relationship of indomethacin analogues for MRP-1, COX-1 and COX-2 inhibition. identification of novel chemotherapeutic drug resistance modulators. Eur. J. Cancer. 2002;38:1661–1670. doi: 10.1016/s0959-8049(02)00128-4. [DOI] [PubMed] [Google Scholar]
- 100.Guo YC, et al. Indomethacin inhibits cancer cell migration via attenuation of cellular calcium mobilization. Molecules. 2013;18:6584–6596. doi: 10.3390/molecules18066584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang YJ, et al. mTOR signaling is involved in indomethacin and nimesulide suppression of colorectal cancer cell growth via a COX-2 independent pathway. Ann. Surg. Oncol. 2011;18:580–588. doi: 10.1245/s10434-010-1268-9. [DOI] [PubMed] [Google Scholar]
- 102.Brunelli C, et al. The non-steroidal anti-inflammatory drug indomethacin activates the eIF2alpha kinase PKR, causing a translational block in human colorectal cancer cells. Biochem J. 2012;443:379–386. doi: 10.1042/BJ20111236. [DOI] [PubMed] [Google Scholar]
- 103.Cheng YL, Zhang GY, Li C, Lin J. Screening for novel protein targets of indomethacin in HCT116 human colon cancer cells using proteomics. Oncol. Lett. 2013;6:1222–1228. doi: 10.3892/ol.2013.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin CC, et al. Targeting the Shc-EGFR interaction with indomethacin inhibits MAP kinase pathway signalling. Cancer Lett. 2019;457:86–97. doi: 10.1016/j.canlet.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mazumder S, et al. Indomethacin impairs mitochondrial dynamics by activating the PKCzeta-p38-DRP1 pathway and inducing apoptosis in gastric cancer and normal mucosal cells. J. Biol. Chem. 2019;294:8238–8258. doi: 10.1074/jbc.RA118.004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cuzick J. Preventive therapy for cancer. Lancet Oncol. 2017;18:e472–e482. doi: 10.1016/S1470-2045(17)30536-3. [DOI] [PubMed] [Google Scholar]
- 107.Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nat. Rev. Clin. Oncol. 2012;9:259–267. doi: 10.1038/nrclinonc.2011.199. [DOI] [PubMed] [Google Scholar]
- 108.Rostom A, et al. Nonsteroidal anti-inflammatory drugs and cyclooxygenase-2 inhibitors for primary prevention of colorectal cancer: a systematic review prepared for the U.S. Preventive Services Task Force. Ann. Intern Med. 2007;146:376–389. doi: 10.7326/0003-4819-146-5-200703060-00010. [DOI] [PubMed] [Google Scholar]
- 109.Nagaraj AB, et al. Using a novel computational drug-repositioning approach (DrugPredict) to rapidly identify potent drug candidates for cancer treatment. Oncogene. 2018;37:403–414. doi: 10.1038/onc.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019;20:175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mohammad RM, et al. Broad targeting of resistance to apoptosis in cancer. Semin Cancer Biol. 2015;35(Suppl):S78–S103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dixon SJ, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smithuis F, et al. Effectiveness of five artemisinin combination regimens with or without primaquine in uncomplicated falciparum malaria: an open-label randomised trial. Lancet Infect. Dis. 2010;10:673–681. doi: 10.1016/S1473-3099(10)70187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ansari MT, et al. Malaria and artemisinin derivatives: an updated review. Mini Rev. Med Chem. 2013;13:1879–1902. doi: 10.2174/13895575113136660097. [DOI] [PubMed] [Google Scholar]
- 115.Karunajeewa HA, et al. Rectal administration of artemisinin derivatives for the treatment of malaria. JAMA. 2007;297:2381–2390. doi: 10.1001/jama.297.21.2381. [DOI] [PubMed] [Google Scholar]
- 116.Mulenga M, Manyando C. Mounting evidence for use of artemisinin derivatives for malaria in early pregnancy. Lancet Infect. Dis. 2016;16:513–515. doi: 10.1016/S1473-3099(16)00019-0. [DOI] [PubMed] [Google Scholar]
- 117.Carter MD, et al. Lipophilic mediated assays for beta-hematin inhibitors. Comb. Chem. High. Throughput Screen. 2010;13:285–292. doi: 10.2174/138620710790980496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ginsburg H, Atamna H. The redox status of malaria-infected erythrocytes: an overview with an emphasis on unresolved problems. Parasite. 1994;1:5–13. doi: 10.1051/parasite/1994011005. [DOI] [PubMed] [Google Scholar]
- 119.Shenai BR, Sijwali PS, Singh A, Rosenthal PJ. Characterization of native and recombinant falcipain-2, a principal trophozoite cysteine protease and essential hemoglobinase of Plasmodium falciparum. J. Biol. Chem. 2000;275:29000–29010. doi: 10.1074/jbc.M004459200. [DOI] [PubMed] [Google Scholar]
- 120.Cumming JN, Ploypradith P, Posner GH. Antimalarial activity of artemisinin (qinghaosu) and related trioxanes: mechanism(s) of action. Adv. Pharmacol. 1997;37:253–297. doi: 10.1016/s1054-3589(08)60952-7. [DOI] [PubMed] [Google Scholar]
- 121.Li WD, Dong YJ, Tu YY, Lin ZB. Dihydroarteannuin ameliorates lupus symptom of BXSB mice by inhibiting production of TNF-alpha and blocking the signaling pathway NF-kappa B translocation. Int Immunopharmacol. 2006;6:1243–1250. doi: 10.1016/j.intimp.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 122.Oiknine-Djian, E. et al. The artemisinin derivative artemisone is a potent inhibitor of human cytomegalovirus replication. Antimicrob. Agents Chemother. 62, (2018). [DOI] [PMC free article] [PubMed]
- 123.Efferth T, et al. The antiviral activities of artemisinin and artesunate. Clin. Infect. Dis. 2008;47:804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 124.Nass J, Efferth T. The activity of Artemisia spp. and their constituents against Trypanosomiasis. Phytomedicine. 2018;47:184–191. doi: 10.1016/j.phymed.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 125.Zhu, S. et al. Ferroptosis: a novel mechanism of artemisinin and its derivatives in cancer therapy. Curr. Med. Chem. 10.2174/0929867327666200121124404 (2020). [DOI] [PubMed]
- 126.Wong YK, et al. Artemisinin as an anticancer drug: recent advances in target profiling and mechanisms of action. Med. Res. Rev. 2017;37:1492–1517. doi: 10.1002/med.21446. [DOI] [PubMed] [Google Scholar]
- 127.Yang ND, et al. Artesunate induces cell death in human cancer cells via enhancing lysosomal function and lysosomal degradation of ferritin. J. Biol. Chem. 2014;289:33425–33441. doi: 10.1074/jbc.M114.564567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen GQ, et al. Artemisinin compounds sensitize cancer cells to ferroptosis by regulating iron homeostasis. Cell Death Differ. 2020;27:242–254. doi: 10.1038/s41418-019-0352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Du J, et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radic. Biol. Med. 2019;131:356–369. doi: 10.1016/j.freeradbiomed.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 130.Liu X, et al. Novel dihydroartemisinin derivative DHA-37 induces autophagic cell death through upregulation of HMGB1 in A549 cells. Cell Death Dis. 2018;9:1048. doi: 10.1038/s41419-018-1006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou X, et al. Artesunate inhibits the growth of gastric cancer cells through the mechanism of promoting oncosis both in vitro and in vivo. Anticancer Drugs. 2013;24:920–927. doi: 10.1097/CAD.0b013e328364a109. [DOI] [PubMed] [Google Scholar]
- 132.Du JH, Zhang HD, Ma ZJ, Ji KM. Artesunate induces oncosis-like cell death in vitro and has antitumor activity against pancreatic cancer xenografts in vivo. Cancer Chemother. Pharmacol. 2010;65:895–902. doi: 10.1007/s00280-009-1095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang J, et al. Dihydroartemisinin induces endothelial cell anoikis through the activation of the JNK signaling pathway. Oncol. Lett. 2016;12:1896–1900. doi: 10.3892/ol.2016.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jansen FH, et al. First study of oral Artenimol-R in advanced cervical cancer: clinical benefit, tolerability and tumor markers. Anticancer Res. 2011;31:4417–4422. [PubMed] [Google Scholar]
- 135.Krishna S, et al. A randomised, double blind, placebo-controlled pilot study of oral artesunate therapy for colorectal cancer. EBioMedicine. 2015;2:82–90. doi: 10.1016/j.ebiom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ericsson T, et al. Effects of artemisinin antimalarials on Cytochrome P450 enzymes in vitro using recombinant enzymes and human liver microsomes: potential implications for combination therapies. Xenobiotica. 2014;44:615–626. doi: 10.3109/00498254.2013.878815. [DOI] [PubMed] [Google Scholar]
- 137.Abarientos C, et al. Hydroxychloroquine in systemic lupus erythematosus and rheumatoid arthritis and its safety in pregnancy. Exp. Opin. Drug Saf. 2011;10:705–714. doi: 10.1517/14740338.2011.566555. [DOI] [PubMed] [Google Scholar]
- 138.Al-Rawi H, Meggitt SJ, Williams FM, Wahie S. Steady-state pharmacokinetics of hydroxychloroquine in patients with cutaneous lupus erythematosus. Lupus. 2018;27:847–852. doi: 10.1177/0961203317727601. [DOI] [PubMed] [Google Scholar]
- 139.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat. Rev. Clin. Oncol. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. [DOI] [PubMed] [Google Scholar]
- 141.Mauthe M, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rebecca VW, et al. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov. 2019;9:220–229. doi: 10.1158/2159-8290.CD-18-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Briceno E, Reyes S, Sotelo J. Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurg. Focus. 2003;14:e3. doi: 10.3171/foc.2003.14.2.4. [DOI] [PubMed] [Google Scholar]
- 144.Vogl DT, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Goldberg SB, et al. A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. J. Thorac. Oncol. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Karasic TB, et al. Effect of gemcitabine and nab-paclitaxel with or without hydroxychloroquine on patients with advanced pancreatic cancer: a Phase 2 randomized clinical trial. JAMA Oncol. 2019;5:993–998. doi: 10.1001/jamaoncol.2019.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wolpin BM, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19:637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chi MS, et al. Double autophagy modulators reduce 2-deoxyglucose uptake in sarcoma patients. Oncotarget. 2015;6:29808–29817. doi: 10.18632/oncotarget.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 151.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 152.Martinez-Outschoorn UE, et al. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 153.Stine ZE, et al. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Desbats MA, Giacomini I, Prayer-Galetti T, Montopoli M. Metabolic plasticity in chemotherapy resistance. Front Oncol. 2020;10:281. doi: 10.3389/fonc.2020.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys. Acta Rev. Cancer. 2017;1868:1–6. doi: 10.1016/j.bbcan.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 156.Pernicova I, Korbonits M. Metformin-mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014;10:143–156. doi: 10.1038/nrendo.2013.256. [DOI] [PubMed] [Google Scholar]
- 157.Brown JB, Nichols GA, Perry A. The burden of treatment failure in type 2 diabetes. Diabetes Care. 2004;27:1535–1540. doi: 10.2337/diacare.27.7.1535. [DOI] [PubMed] [Google Scholar]
- 158.Rao Kondapally Seshasai S, et al. Diabetes mellitus, fasting glucose, and risk of cause-specific death. N. Engl. J. Med. 2011;364:829–841. doi: 10.1056/NEJMoa1008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peairs KS, et al. Diabetes mellitus and breast cancer outcomes: a systematic review and meta-analysis. J. Clin. Oncol. 2011;29:40–46. doi: 10.1200/JCO.2009.27.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gallagher EJ, LeRoith D. Obesity and diabetes: the increased risk of cancer and cancer-related mortality. Physiol. Rev. 2015;95:727–748. doi: 10.1152/physrev.00030.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pollak M. Overcoming drug development bottlenecks with repurposing: repurposing biguanides to target energy metabolism for cancer treatment. Nat. Med. 2014;20:591–593. doi: 10.1038/nm.3596. [DOI] [PubMed] [Google Scholar]
- 162.Bronsveld HK, et al. The association of diabetes mellitus and insulin treatment with expression of insulin-related proteins in breast tumors. BMC Cancer. 2018;18:224. doi: 10.1186/s12885-018-4072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Howell JJ, et al. Metformin inhibits hepatic mTORC1 signaling via dose-dependent mechanisms involving AMPK and the TSC complex. Cell Metab. 2017;25:463–471. doi: 10.1016/j.cmet.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kalender A, et al. Metformin, independent of AMPK, inhibits mTORC1 in a RAG GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu D, et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature. 2018;559:637–641. doi: 10.1038/s41586-018-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu X, et al. Metformin targets central carbon metabolism and reveals mitochondrial requirements in human cancers. Cell Metab. 2016;24:728–739. doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lee J, et al. Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature. 2019;568:254–258. doi: 10.1038/s41586-019-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mark M, et al. Impact of addition of metformin to abiraterone in metastatic castration-resistant prostate cancer patients with disease progressing while receiving abiraterone treatment (MetAb-Pro): phase 2 pilot study. Clin. Genitourin. Cancer. 2019;17:e323–e328. doi: 10.1016/j.clgc.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 169.Zhang ZJ, et al. The effect of metformin on biomarkers and survivals for breast cancer- a systematic review and meta-analysis of randomized clinical trials. Pharm. Res. 2019;141:551–555. doi: 10.1016/j.phrs.2019.01.036. [DOI] [PubMed] [Google Scholar]
- 170.Petchsila K, et al. Effect of metformin for decreasing proliferative marker in women with endometrial cancer: a randomized double-blind placebo-controlled trial. Asian Pac. J. Cancer Prev. 2020;21:733–741. doi: 10.31557/APJCP.2020.21.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bhaw-Luximon A, Jhurry D. Metformin in pancreatic cancer treatment: from clinical trials through basic research to biomarker quantification. J. Cancer Res Clin. Oncol. 2016;142:2159–2171. doi: 10.1007/s00432-016-2178-4. [DOI] [PubMed] [Google Scholar]
- 172.Hald J, Jacobsen E. A drug sensitizing the organism to ethyl alcohol. Lancet. 1948;2:1001–1004. doi: 10.1016/s0140-6736(48)91514-1. [DOI] [PubMed] [Google Scholar]
- 173.Suh JJ, Pettinati HM, Kampman KM, O'Brien CP. The status of disulfiram: a half of a century later. J. Clin. Psychopharmacol. 2006;26:290–302. doi: 10.1097/01.jcp.0000222512.25649.08. [DOI] [PubMed] [Google Scholar]
- 174.Lipsky JJ, Shen ML, Naylor S. Overview-in vitro inhibition of aldehyde dehydrogenase by disulfiram and metabolites. Chem. Biol. Interact. 2001;130–132:81–91. doi: 10.1016/s0009-2797(00)00224-6. [DOI] [PubMed] [Google Scholar]
- 175.Veverka KA, et al. Inhibition of aldehyde dehydrogenase by disulfiram and its metabolite methyl diethylthiocarbamoyl-sulfoxide. Biochem. Pharmacol. 1997;53:511–518. doi: 10.1016/s0006-2952(96)00767-8. [DOI] [PubMed] [Google Scholar]
- 176.Ekinci E, Rohondia S, Khan R, Dou QP. Repurposing disulfiram as an anticancer agent: updated review on literature and patents. Recent Pat. Anticancer Drug Discov. 2019;14:113–132. doi: 10.2174/1574892814666190514104035. [DOI] [PubMed] [Google Scholar]
- 177.Tacconi EM, et al. BRCA1 and BRCA2 tumor suppressors protect against endogenous acetaldehyde toxicity. EMBO Mol. Med. 2017;9:1398–1414. doi: 10.15252/emmm.201607446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Choi SA, et al. Disulfiram modulates stemness and metabolism of brain tumor initiating cells in atypical teratoid/rhabdoid tumors. Neuro Oncol. 2015;17:810–821. doi: 10.1093/neuonc/nou305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dorokhov YL, Sheshukova EV, Bialik TE, Komarova TV. Human endogenous formaldehyde as an anticancer metabolite: its oxidation downregulation may be a means of improving therapy. Bioessays. 2018;40:e1800136. doi: 10.1002/bies.201800136. [DOI] [PubMed] [Google Scholar]
- 180.Pontel LB, et al. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015;60:177–188. doi: 10.1016/j.molcel.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guo F, et al. Inhibitory effect on ovarian cancer ALDH+ stem-like cells by disulfiram and copper treatment through ALDH and ROS modulation. Biomed. Pharmacother. 2019;118:109371. doi: 10.1016/j.biopha.2019.109371. [DOI] [PubMed] [Google Scholar]
- 182.Liu P, et al. Cytotoxic effect of disulfiram/copper on human glioblastoma cell lines and ALDH-positive cancer-stem-like cells. Br. J. Cancer. 2012;107:1488–1497. doi: 10.1038/bjc.2012.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cong J, et al. A novel chemoradiation targeting stem and nonstem pancreatic cancer cells by repurposing disulfiram. Cancer Lett. 2017;409:9–19. doi: 10.1016/j.canlet.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 184.Shian SG, Kao YR, Wu FY, Wu CW. Inhibition of invasion and angiogenesis by zinc-chelating agent disulfiram. Mol. Pharmacol. 2003;64:1076–1084. doi: 10.1124/mol.64.5.1076. [DOI] [PubMed] [Google Scholar]
- 185.Falls-Hubert KC, et al. Disulfiram causes selective hypoxic cancer cell toxicity and radio-chemo-sensitization via redox cycling of copper. Free Radic. Biol. Med. 2020;150:1–11. doi: 10.1016/j.freeradbiomed.2020.01.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sonawane VK, et al. A chemosensitizer drug: disulfiram prevents doxorubicin-induced cardiac dysfunction and oxidative stress in rats. Cardiovasc. Toxicol. 2018;18:459–470. doi: 10.1007/s12012-018-9458-y. [DOI] [PubMed] [Google Scholar]
- 187.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells—mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 2017;14:155–167. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 188.Teng MW, et al. Immune-mediated dormancy: an equilibrium with cancer. J. Leukoc. Biol. 2008;84:988–993. doi: 10.1189/jlb.1107774. [DOI] [PubMed] [Google Scholar]
- 189.Dunn GP, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 190.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 191.Mulder WJM, et al. Therapeutic targeting of trained immunity. Nat. Rev. Drug Discov. 2019;18:553–566. doi: 10.1038/s41573-019-0025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rappuoli R, Mandl CW, Black S, De Gregorio E. Vaccines for the twenty-first century society. Nat. Rev. Immunol. 2011;11:865–872. doi: 10.1038/nri3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Melero IP, et al. Repurposing infectious disease vaccines for intratumoral immunotherapy. J. Immunother. Cancer. 2020;8:e000443. doi: 10.1136/jitc-2019-000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shekarian T, et al. Repurposing rotavirus vaccines for intratumoral immunotherapy can overcome resistance to immune checkpoint blockade. Sci. Transl. Med. 2019;11:eaat5025. doi: 10.1126/scitranslmed.aat5025. [DOI] [PubMed] [Google Scholar]
- 195.Aznar MA, et al. Repurposing the yellow fever vaccine for intratumoral immunotherapy. EMBO Mol. Med. 2020;12:e10375. doi: 10.15252/emmm.201910375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mastrangelo G, et al. Is there a relationship between influenza vaccinations and risk of melanoma? A population-based case-control study. Eur. J. Epidemiol. 2000;16:777–782. doi: 10.1023/a:1007658503740. [DOI] [PubMed] [Google Scholar]
- 197.Sun W, Sanderson PE, Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov. Today. 2016;21:1189–1195. doi: 10.1016/j.drudis.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell. 2017;171:1678–1691. doi: 10.1016/j.cell.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tyers M, Wright GD. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019;17:141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- 201.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 202.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 203.Williams BO, et al. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat. Genet. 1994;7:480–484. doi: 10.1038/ng0894-480. [DOI] [PubMed] [Google Scholar]
- 204.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 206.Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nat. Rev. Cancer. 2012;12:220–226. doi: 10.1038/nrc3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Napoli M, Flores ER. Loss of p53 protein strikes a nerve to aid tumour growth. Nature. 2020;578:367–369. doi: 10.1038/d41586-020-00328-6. [DOI] [PubMed] [Google Scholar]
- 208.Xu XL, et al. Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature. 2014;514:385–388. doi: 10.1038/nature13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Van Dyke K, Lantz C, Szustkiewicz C. Quinacrine: mechanisms of antimalarial action. Science. 1970;169:492–493. [PubMed] [Google Scholar]
- 210.Requena-Mendez A, et al. The use of quinacrine in nitroimidazole-resistant giardia duodenalis: an old drug for an emerging problem. J. Infect. Dis. 2017;215:946–953. doi: 10.1093/infdis/jix066. [DOI] [PubMed] [Google Scholar]
- 211.Egorin MJ, Trump DL, Wainwright CW. Quinacrine ochronosis and rheumatoid arthritis. JAMA. 1976;236:385–386. [PubMed] [Google Scholar]
- 212.Larrieu AJ, et al. Intrapleural instillation of quinacrine for treatment of recurrent spontaneous pneumothorax. Ann. Thorac. Surg. 1979;28:146–150. doi: 10.1016/s0003-4975(10)63772-1. [DOI] [PubMed] [Google Scholar]
- 213.Geschwind MD, et al. Quinacrine treatment trial for sporadic Creutzfeldt-Jakob disease. Neurology. 2013;81:2015–2023. doi: 10.1212/WNL.0b013e3182a9f3b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Collinge J, et al. Safety and efficacy of quinacrine in human prion disease (PRION-1 study): a patient-preference trial. Lancet Neurol. 2009;8:334–344. doi: 10.1016/S1474-4422(09)70049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Oien, D. B. et al. Repurposing quinacrine for treatment-refractory cancer. Semin. Cancer Biol. 10.1016/j.semcancer.2019.09.021 (2019). [DOI] [PubMed]
- 216.Gurova KV, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc. Natl Acad. Sci. USA. 2005;102:17448–17453. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nesher E, et al. Role of chromatin damage and chromatin trapping of FACT in mediating the anticancer cytotoxicity of DNA-binding small-molecule drugs. Cancer Res. 2018;78:1431–1443. doi: 10.1158/0008-5472.CAN-17-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gasparian AV, et al. Curaxins: anticancer compounds that simultaneously suppress NF-kappaB and activate p53 by targeting FACT. Sci. Transl. Med. 2011;3:95ra74. doi: 10.1126/scitranslmed.3002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Park S, et al. Therapeutic effect of quinacrine, an antiprotozoan drug, by selective suppression of p-CHK1/2 in p53-negative malignant cancers. Mol. Cancer Res. 2018;16:935–946. doi: 10.1158/1541-7786.MCR-17-0511. [DOI] [PubMed] [Google Scholar]
- 220.Preet R, et al. Chk1 inhibitor synergizes quinacrine mediated apoptosis in breast cancer cells by compromising the base excision repair cascade. Biochem. Pharmacol. 2016;105:23–33. doi: 10.1016/j.bcp.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 221.Mohapatra P, et al. Quinacrine-mediated autophagy and apoptosis in colon cancer cells is through a p53- and p21-dependent mechanism. Oncol. Res. 2012;20:81–91. doi: 10.3727/096504012x13473664562628. [DOI] [PubMed] [Google Scholar]
- 222.Hede K. Antimalaria drug offers antitumor strategies. J. Natl Cancer Inst. 2011;103:1490–1491. doi: 10.1093/jnci/djr423. [DOI] [PubMed] [Google Scholar]
- 223.Markowitz M, et al. A preliminary study of ritonavir, an inhibitor of HIV-1 protease, to treat HIV-1 infection. N. Engl. J. Med. 1995;333:1534–1539. doi: 10.1056/NEJM199512073332204. [DOI] [PubMed] [Google Scholar]
- 224.Gallant JE. Strategies for long-term success in the treatment of HIV infection. JAMA. 2000;283:1329–1334. doi: 10.1001/jama.283.10.1329. [DOI] [PubMed] [Google Scholar]
- 225.Simon V, Ho DD, Abdool Karim Q. HIV/AIDS epidemiology, pathogenesis, prevention, and treatment. Lancet. 2006;368:489–504. doi: 10.1016/S0140-6736(06)69157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Eggleton, J. S. & Nagalli, S. in StatPearls (2020).
- 227.Franceschi S, et al. Kaposi sarcoma incidence in the Swiss HIV Cohort Study before and after highly active antiretroviral therapy. Br. J. Cancer. 2008;99:800–804. doi: 10.1038/sj.bjc.6604520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Franceschi S, et al. Changing patterns of cancer incidence in the early- and late-HAART periods: the Swiss HIV Cohort Study. Br. J. Cancer. 2010;103:416–422. doi: 10.1038/sj.bjc.6605756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rengan R, et al. Effect of HIV on survival in patients with non-small-cell lung cancer in the era of highly active antiretroviral therapy: a population-based study. Lancet Oncol. 2012;13:1203–1209. doi: 10.1016/S1470-2045(12)70466-7. [DOI] [PubMed] [Google Scholar]
- 230.Lim ST, et al. AIDS-related Burkitt's lymphoma versus diffuse large-cell lymphoma in the pre-highly active antiretroviral therapy (HAART) and HAART eras: significant differences in survival with standard chemotherapy. J. Clin. Oncol. 2005;23:4430–4438. doi: 10.1200/JCO.2005.11.973. [DOI] [PubMed] [Google Scholar]
- 231.Chiao EY, Giordano TP, Richardson P, El-Serag HB. Human immunodeficiency virus-associated squamous cell cancer of the anus: epidemiology and outcomes in the highly active antiretroviral therapy era. J. Clin. Oncol. 2008;26:474–479. doi: 10.1200/JCO.2007.14.2810. [DOI] [PubMed] [Google Scholar]
- 232.Chow WA, Jiang C, Guan M. Anti-HIV drugs for cancer therapeutics: back to the future? Lancet Oncol. 2009;10:61–71. doi: 10.1016/S1470-2045(08)70334-6. [DOI] [PubMed] [Google Scholar]
- 233.Gaedicke S, et al. Antitumor effect of the human immunodeficiency virus protease inhibitor ritonavir: induction of tumor-cell apoptosis associated with perturbation of proteasomal proteolysis. Cancer Res. 2002;62:6901–6908. [PubMed] [Google Scholar]
- 234.Kumar S, et al. Ritonavir blocks AKT signaling, activates apoptosis and inhibits migration and invasion in ovarian cancer cells. Mol. Cancer. 2009;8:26. doi: 10.1186/1476-4598-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Srirangam A, et al. The human immunodeficiency virus protease inhibitor ritonavir inhibits lung cancer cells, in part, by inhibition of survivin. J. Thorac. Oncol. 2011;6:661–670. doi: 10.1097/JTO.0b013e31820c9e3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Batchu RB, et al. Ritonavir-mediated induction of apoptosis in pancreatic cancer occurs via the RB/E2F-1 and AKT pathways. Pharmacology. 2014;7:46–57. doi: 10.3390/ph7010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ahluwalia MS, et al. Phase II trial of ritonavir/lopinavir in patients with progressive or recurrent high-grade gliomas. J. Neurooncol. 2011;102:317–321. doi: 10.1007/s11060-010-0325-3. [DOI] [PubMed] [Google Scholar]
- 238.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 239.Schmitt CA. Senescence, apoptosis and therapy-cutting the lifelines of cancer. Nat. Rev. Cancer. 2003;3:286–295. doi: 10.1038/nrc1044. [DOI] [PubMed] [Google Scholar]
- 240.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Arndt GM, MacKenzie KL. New prospects for targeting telomerase beyond the telomere. Nat. Rev. Cancer. 2016;16:508–524. doi: 10.1038/nrc.2016.55. [DOI] [PubMed] [Google Scholar]
- 242.Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 2019;20:299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 243.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shkreli M, et al. Reversible cell-cycle entry in adult kidney podocytes through regulated control of telomerase and Wnt signaling. Nat. Med. 2011;18:111–119. doi: 10.1038/nm.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hoffmeyer K, et al. Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science. 2012;336:1549–1554. doi: 10.1126/science.1218370. [DOI] [PubMed] [Google Scholar]
- 246.Varelas X, Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends Cell Biol. 2012;22:88–96. doi: 10.1016/j.tcb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 247.Vermeulen L. Keeping stem cells in check: a hippo balancing act. Cell Stem Cell. 2013;12:3–5. doi: 10.1016/j.stem.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 248.Kim W, et al. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. J. Clin. Investig. 2017;127:137–152. doi: 10.1172/JCI88486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Vaughn AR, Branum A, Sivamani RK. Effects of turmeric (Curcuma longa) on skin health: a systematic review of the clinical evidence. Phytother. Res. 2016;30:1243–1264. doi: 10.1002/ptr.5640. [DOI] [PubMed] [Google Scholar]
- 250.Fowler JF, Jr, Woolery-Lloyd H, Waldorf H, Saini R. Innovations in natural ingredients and their use in skin care. J. Drugs Dermatol. 2010;9:S72–S81. [PubMed] [Google Scholar]
- 251.Wanninger S, Lorenz V, Subhan A, Edelmann FT. Metal complexes of curcumin-synthetic strategies, structures and medicinal applications. Chem. Soc. Rev. 2015;44:4986–5002. doi: 10.1039/c5cs00088b. [DOI] [PubMed] [Google Scholar]
- 252.Patel SS, et al. Cellular and molecular mechanisms of curcumin in prevention and treatment of disease. Crit. Rev. Food Sci. Nutr. 2020;60:887–939. doi: 10.1080/10408398.2018.1552244. [DOI] [PubMed] [Google Scholar]
- 253.Ramachandran C, et al. Curcumin inhibits telomerase activity through human telomerase reverse transcritpase in MCF-7 breast cancer cell line. Cancer Lett. 2002;184:1–6. doi: 10.1016/s0304-3835(02)00192-1. [DOI] [PubMed] [Google Scholar]
- 254.Nasiri M, et al. Curcumin and silibinin inhibit telomerase expression in T47D human breast cancer cells. Asian Pac. J. Cancer Prev. 2013;14:3449–3453. doi: 10.7314/apjcp.2013.14.6.3449. [DOI] [PubMed] [Google Scholar]
- 255.Kazemi-Lomedasht F, Rami A, Zarghami N. Comparison of inhibitory effect of curcumin nanoparticles and free curcumin in human telomerase reverse transcriptase gene expression in breast cancer. Adv. Pharm. Bull. 2013;3:127–130. doi: 10.5681/apb.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lee JH, Chung IK. Curcumin inhibits nuclear localization of telomerase by dissociating the Hsp90 co-chaperone p23 from hTERT. Cancer Lett. 2010;290:76–86. doi: 10.1016/j.canlet.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 257.Aravindan N, et al. Curcumin regulates low-linear energy transfer gamma-radiation-induced NFkappa B-dependent telomerase activity in human neuroblastoma cells. Int J. Radiat. Oncol. Biol. Phys. 2011;79:1206–1215. doi: 10.1016/j.ijrobp.2010.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Chung SS, et al. A novel curcumin analog inhibits canonical and non-canonical functions of telomerase through STAT3 and NF-kappaB inactivation in colorectal cancer cells. Oncotarget. 2019;10:4516–4531. doi: 10.18632/oncotarget.27000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cianciosi D, et al. Targeting molecular pathways in cancer stem cells by natural bioactive compounds. Pharm. Res. 2018;135:150–165. doi: 10.1016/j.phrs.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 260.Liang Z, et al. Curcumin reversed chronic tobacco smoke exposure induced urocystic EMT and acquisition of cancer stem cells properties via Wnt/beta-catenin. Cell Death Dis. 2017;8:e3066. doi: 10.1038/cddis.2017.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Li Y, Zhang T. Targeting cancer stem cells by curcumin and clinical applications. Cancer Lett. 2014;346:197–205. doi: 10.1016/j.canlet.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 262.Sarkar FH, Li Y, Wang Z, Kong D. The role of nutraceuticals in the regulation of Wnt and Hedgehog signaling in cancer. Cancer Metastasis Rev. 2010;29:383–394. doi: 10.1007/s10555-010-9233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ye C, et al. A novel curcumin derivative CL-6 exerts antitumor effect in human gastric cancer cells by inducing apoptosis through Hippo-YAP signaling pathway. Onco Targets Ther. 2019;12:2259–2269. doi: 10.2147/OTT.S196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gao Y, et al. Curcumin promotes KLF5 proteasome degradation through downregulating YAP/TAZ in bladder cancer cells. Int J. Mol. Sci. 2014;15:15173–15187. doi: 10.3390/ijms150915173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Shen Y, et al. Curcumin inhibits the tumorigenesis of breast cancer by blocking tafazzin/yes-associated protein axis. Cancer Manag. Res. 2020;12:1493–1502. doi: 10.2147/CMAR.S246691. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 266.Kunnumakkara AB, et al. Is curcumin bioavailability a problem in humans: lessons from clinical trials. Exp. Opin. Drug Metab. Toxicol. 2019;15:705–733. doi: 10.1080/17425255.2019.1650914. [DOI] [PubMed] [Google Scholar]
- 267.Dei Cas M, Ghidoni R. Dietary curcumin: correlation between bioavailability and health potential. Nutrients. 2019;11:2147. doi: 10.3390/nu11092147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Heger M, van Golen RF, Broekgaarden M, Michel MC. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharm. Rev. 2014;66:222–307. doi: 10.1124/pr.110.004044. [DOI] [PubMed] [Google Scholar]
- 269.Kanai M. Therapeutic applications of curcumin for patients with pancreatic cancer. World J. Gastroenterol. 2014;20:9384–9391. doi: 10.3748/wjg.v20.i28.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hosseini M, et al. Therapeutic potential of curcumin in treatment of pancreatic cancer: current status and future perspectives. J. Cell Biochem. 2017;118:1634–1638. doi: 10.1002/jcb.25897. [DOI] [PubMed] [Google Scholar]
- 271.Thangavel P, Puga-Olguin A, Rodriguez-Landa JF, Zepeda RC. Genistein as potential therapeutic candidate for menopausal symptoms and other related diseases. Molecules. 2019;24:3892. doi: 10.3390/molecules24213892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Cotter A, Cashman KD. Genistein appears to prevent early postmenopausal bone loss as effectively as hormone replacement therapy. Nutr. Rev. 2003;61:346–351. doi: 10.1301/nr.2003.oct.346-351. [DOI] [PubMed] [Google Scholar]
- 273.Dang ZC. Dose-dependent effects of soy phyto-oestrogen genistein on adipocytes: mechanisms of action. Obes. Rev. 2009;10:342–349. doi: 10.1111/j.1467-789X.2008.00554.x. [DOI] [PubMed] [Google Scholar]
- 274.Verheus M, et al. Plasma phytoestrogens and subsequent breast cancer risk. J. Clin. Oncol. 2007;25:648–655. doi: 10.1200/JCO.2006.06.0244. [DOI] [PubMed] [Google Scholar]
- 275.Gescher AJ, Sharma RA, Steward WP. Cancer chemoprevention by dietary constituents: a tale of failure and promise. Lancet Oncol. 2001;2:371–379. doi: 10.1016/S1470-2045(00)00392-2. [DOI] [PubMed] [Google Scholar]
- 276.Spagnuolo C, et al. Genistein and cancer: current status, challenges, and future directions. Adv. Nutr. 2015;6:408–419. doi: 10.3945/an.114.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ouchi H, et al. Genistein induces cell growth inhibition in prostate cancer through the suppression of telomerase activity. Int. J. Urol. 2005;12:73–80. doi: 10.1111/j.1442-2042.2004.00973.x. [DOI] [PubMed] [Google Scholar]
- 278.Jagadeesh S, Kyo S, Banerjee PP. Genistein represses telomerase activity via both transcriptional and posttranslational mechanisms in human prostate cancer cells. Cancer Res. 2006;66:2107–2115. doi: 10.1158/0008-5472.CAN-05-2494. [DOI] [PubMed] [Google Scholar]
- 279.Khaw AK, Yong JW, Kalthur G, Hande MP. Genistein induces growth arrest and suppresses telomerase activity in brain tumor cells. Genes Chromosomes Cancer. 2012;51:961–974. doi: 10.1002/gcc.21979. [DOI] [PubMed] [Google Scholar]
- 280.Alhasan SA, Aranha O, Sarkar FH. Genistein elicits pleiotropic molecular effects on head and neck cancer cells. Clin. Cancer Res. 2001;7:4174–4181. [PubMed] [Google Scholar]
- 281.Ozturk SA, et al. The effects of thymoquinone and genistein treatment on telomerase activity, apoptosis, angiogenesis, and survival in thyroid cancer cell lines. J. Cancer Res. Ther. 2018;14:328–334. doi: 10.4103/0973-1482.202886. [DOI] [PubMed] [Google Scholar]
- 282.Li Y, Liu L, Andrews LG, Tollefsbol TO. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int. J. Cancer. 2009;125:286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Hirata H, et al. Genistein downregulates onco-miR-1260b and inhibits Wnt-signalling in renal cancer cells. Br. J. Cancer. 2013;108:2070–2078. doi: 10.1038/bjc.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zhang Y, Li Q, Chen H. DNA methylation and histone modifications of Wnt genes by genistein during colon cancer development. Carcinogenesis. 2013;34:1756–1763. doi: 10.1093/carcin/bgt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Zhang Y, Chen H. Genistein attenuates WNT signaling by up-regulating sFRP2 in a human colon cancer cell line. Exp. Biol. Med. 2011;236:714–722. doi: 10.1258/ebm.2011.010347. [DOI] [PubMed] [Google Scholar]
- 286.Naujokat, C. & McKee, D. L. The “Big Five” phytochemicals targeting cancer stem cells: curcumin, EGCG, sulforaphane, resveratrol and genistein. Curr. Med. Chem. 10.2174/0929867327666200228110738 (2020). [DOI] [PubMed]
- 287.Amin A, et al. Evasion of anti-growth signaling: a key step in tumorigenesis and potential target for treatment and prophylaxis by natural compounds. Semin. Cancer Biol. 2015;35(Suppl):S55–S77. doi: 10.1016/j.semcancer.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Shen H, et al. Preparation, characterization, and pharmacokinetics study of a novel genistein-loaded mixed micelles system. Drug Dev. Ind. Pharm. 2018;44:1536–1542. doi: 10.1080/03639045.2018.1483384. [DOI] [PubMed] [Google Scholar]
- 289.Albini A, et al. Cancer prevention by targeting angiogenesis. Nat. Rev. Clin. Oncol. 2012;9:498–509. doi: 10.1038/nrclinonc.2012.120. [DOI] [PubMed] [Google Scholar]
- 290.De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer. 2017;17:457–474. doi: 10.1038/nrc.2017.51. [DOI] [PubMed] [Google Scholar]
- 291.Rafii S, Lyden D. Cancer. A few to flip the angiogenic switch. Science. 2008;319:163–164. doi: 10.1126/science.1153615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 293.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 294.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lary JM, et al. The return of thalidomide: can birth defects be prevented? Drug Saf. 1999;21:161–169. doi: 10.2165/00002018-199921030-00002. [DOI] [PubMed] [Google Scholar]
- 296.Lindner S, Kronke J. The molecular mechanism of thalidomide analogs in hematologic malignancies. J. Mol. Med. 2016;94:1327–1334. doi: 10.1007/s00109-016-1450-z. [DOI] [PubMed] [Google Scholar]
- 297.Diamanti A, et al. The clinical implications of thalidomide in inflammatory bowel diseases. Exp. Rev. Clin. Immunol. 2015;11:699–708. doi: 10.1586/1744666X.2015.1027687. [DOI] [PubMed] [Google Scholar]
- 298.Liu T, et al. Thalidomide and its analogues: a review of the potential for immunomodulation of fibrosis diseases and opthalmopathy. Exp. Ther. Med. 2017;14:5251–5257. doi: 10.3892/etm.2017.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Richardson P, Anderson K. Thalidomide and dexamethasone: a new standard of care for initial therapy in multiple myeloma. J. Clin. Oncol. 2006;24:334–336. doi: 10.1200/JCO.2005.03.8851. [DOI] [PubMed] [Google Scholar]
- 300.Eleutherakis-Papaiakovou V, Bamias A, Dimopoulos MA. Thalidomide in cancer medicine. Ann. Oncol. 2004;15:1151–1160. doi: 10.1093/annonc/mdh300. [DOI] [PubMed] [Google Scholar]
- 301.Shenoy SK, et al. beta-arrestin1 mediates metastatic growth of breast cancer cells by facilitating HIF-1-dependent VEGF expression. Oncogene. 2012;31:282–292. doi: 10.1038/onc.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Yabu T, et al. Thalidomide-induced antiangiogenic action is mediated by ceramide through depletion of VEGF receptors, and is antagonized by sphingosine-1-phosphate. Blood. 2005;106:125–134. doi: 10.1182/blood-2004-09-3679. [DOI] [PubMed] [Google Scholar]
- 303.Yang Y, Ge JP, Zhou ZT. Effects of thalidomide on DMBA-induced oral carcinogenesis in hamster with respect to angiogenesis. J. Oral. Pathol. Med. 2009;38:455–462. doi: 10.1111/j.1600-0714.2008.00737.x. [DOI] [PubMed] [Google Scholar]
- 304.Li M, et al. In vitro anticancer property of a novel thalidomide analogue through inhibition of NF-kappaB activation in HL-60 cells. Acta Pharm. Sin. 2009;30:134–140. doi: 10.1038/aps.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Huang YT, Cheng CC, Chiu TH, Lai PC. Therapeutic potential of thalidomide for gemcitabine-resistant bladder cancer. Int J. Oncol. 2015;47:1711–1724. doi: 10.3892/ijo.2015.3155. [DOI] [PubMed] [Google Scholar]
- 306.Adlard JW. Thalidomide in the treatment of cancer. Anticancer Drugs. 2000;11:787–791. doi: 10.1097/00001813-200011000-00002. [DOI] [PubMed] [Google Scholar]
- 307.Figg WD, et al. A double-blind randomized crossover study of oral thalidomide versus placebo for androgen dependent prostate cancer treated with intermittent androgen ablation. J. Urol. 2009;181:1104–1113. doi: 10.1016/j.juro.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Como JA, Dismukes WE. Oral azole drugs as systemic antifungal therapy. N. Engl. J. Med. 1994;330:263–272. doi: 10.1056/NEJM199401273300407. [DOI] [PubMed] [Google Scholar]
- 309.Rudin CM, et al. Phase 2 study of pemetrexed and itraconazole as second-line therapy for metastatic nonsquamous non-small-cell lung cancer. J. Thorac. Oncol. 2013;8:619–623. doi: 10.1097/JTO.0b013e31828c3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kim DJ, et al. Open-label, exploratory phase II trial of oral itraconazole for the treatment of basal cell carcinoma. J. Clin. Oncol. 2014;32:745–751. doi: 10.1200/JCO.2013.49.9525. [DOI] [PubMed] [Google Scholar]
- 311.Kim J, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17:388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Tsubamoto H, et al. Repurposing itraconazole as an anticancer agent. Oncol. Lett. 2017;14:1240–1246. doi: 10.3892/ol.2017.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Pounds R, Leonard S, Dawson C, Kehoe S. Repurposing itraconazole for the treatment of cancer. Oncol. Lett. 2017;14:2587–2597. doi: 10.3892/ol.2017.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Antonarakis ES, et al. Repurposing itraconazole as a treatment for advanced prostate cancer: a noncomparative randomized phase II trial in men with metastatic castration-resistant prostate cancer. Oncologist. 2013;18:163–173. doi: 10.1634/theoncologist.2012-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Chong CR, et al. Inhibition of angiogenesis by the antifungal drug itraconazole. ACS Chem. Biol. 2007;2:263–270. doi: 10.1021/cb600362d. [DOI] [PubMed] [Google Scholar]
- 316.Nacev BA, et al. The antifungal drug itraconazole inhibits vascular endothelial growth factor receptor 2 (VEGFR2) glycosylation, trafficking, and signaling in endothelial cells. J. Biol. Chem. 2011;286:44045–44056. doi: 10.1074/jbc.M111.278754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Head SA, et al. Simultaneous targeting of NPC1 and VDAC1 by itraconazole leads to synergistic inhibition of mTOR signaling and angiogenesis. ACS Chem. Biol. 2017;12:174–182. doi: 10.1021/acschembio.6b00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Chen S, et al. Itraconazole induces regression of infantile hemangioma via downregulation of the platelet-derived growth factor-D/PI3K/Akt/mTOR pathway. J. Investig. Dermatol. 2019;139:1574–1582. doi: 10.1016/j.jid.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 319.Tsubamoto H, Sonoda T, Yamasaki M, Inoue K. Impact of combination chemotherapy with itraconazole on survival of patients with refractory ovarian cancer. Anticancer Res. 2014;34:2481–2487. [PubMed] [Google Scholar]
- 320.Tsubamoto H, et al. Combination chemotherapy with itraconazole for treating metastatic pancreatic cancer in the second-line or additional setting. Anticancer Res. 2015;35:4191–4196. [PubMed] [Google Scholar]
- 321.Tsubamoto H, Sonoda T, Inoue K. Impact of itraconazole on the survival of heavily pre-treated patients with triple-negative breast cancer. Anticancer Res. 2014;34:3839–3844. [PubMed] [Google Scholar]
- 322.Copley MS, et al. Itraconazole vs. posaconazole for antifungal prophylaxis in patients with acute myeloid leukemia undergoing intensive chemotherapy: a retrospective study. Int J. Antimicrob. Agents. 2020;55:105886. doi: 10.1016/j.ijantimicag.2020.105886. [DOI] [PubMed] [Google Scholar]
- 323.Ringshausen I, et al. Antifungal therapy with itraconazole impairs the anti-lymphoma effects of rituximab by inhibiting recruitment of CD20 to cell surface lipid rafts. Cancer Res. 2010;70:4292–4296. doi: 10.1158/0008-5472.CAN-10-0259. [DOI] [PubMed] [Google Scholar]
- 324.Yang BR, et al. Co-medication of statins with contraindicated drugs. PLoS ONE. 2015;10:e0125180. doi: 10.1371/journal.pone.0125180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Stuelten CH, Parent CA, Montell DJ. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat. Rev. Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Bandyopadhyay S, et al. Potential antibacterial activity of berberine against multi drug resistant enterovirulent Escherichia coli isolated from yaks (Poephagus grunniens) with haemorrhagic diarrhoea. Asian Pac. J. Trop. Med. 2013;6:315–319. doi: 10.1016/S1995-7645(13)60063-2. [DOI] [PubMed] [Google Scholar]
- 327.Wu N, Sarna LK, Siow YL, O K. Regulation of hepatic cholesterol biosynthesis by berberine during hyperhomocysteinemia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R635–R643. doi: 10.1152/ajpregu.00441.2010. [DOI] [PubMed] [Google Scholar]
- 328.Chang X, et al. Lipid profiling of the therapeutic effects of berberine in patients with nonalcoholic fatty liver disease. J. Transl. Med. 2016;14:266. doi: 10.1186/s12967-016-0982-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Derosa G, D'Angelo A, Vanelli A, Maffioli P. An evaluation of a nutraceutical with berberine, curcumin, inositol, banaba and chromium picolinate in patients with fasting dysglycemia. Diabetes Metab. Syndr. Obes. 2020;13:653–661. doi: 10.2147/DMSO.S232791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Shen N, Huan Y, Shen ZF. Berberine inhibits mouse insulin gene promoter through activation of AMP activated protein kinase and may exert beneficial effect on pancreatic beta-cell. Eur. J. Pharmacol. 2012;694:120–126. doi: 10.1016/j.ejphar.2012.07.052. [DOI] [PubMed] [Google Scholar]
- 331.Zhang H, et al. Berberine lowers blood glucose in type 2 diabetes mellitus patients through increasing insulin receptor expression. Metabolism. 2010;59:285–292. doi: 10.1016/j.metabol.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 332.Ma W, et al. Berberine inhibits the proliferation and migration of breast cancer ZR-75-30 cells by targeting Ephrin-B2. Phytomedicine. 2017;25:45–51. doi: 10.1016/j.phymed.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 333.Liu X, et al. Berberine inhibits invasion and metastasis of colorectal cancer cells via COX-2/PGE2 mediated JAK2/STAT3 signaling pathway. PLoS ONE. 2015;10:e0123478. doi: 10.1371/journal.pone.0123478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kim HS, et al. Berberine-induced AMPK activation inhibits the metastatic potential of melanoma cells via reduction of ERK activity and COX-2 protein expression. Biochem. Pharmacol. 2012;83:385–394. doi: 10.1016/j.bcp.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 335.Wang Y, Zhang S. Berberine suppresses growth and metastasis of endometrial cancer cells via miR-101/COX-2. Biomed. Pharmacother. 2018;103:1287–1293. doi: 10.1016/j.biopha.2018.04.161. [DOI] [PubMed] [Google Scholar]
- 336.Hamsa TP, Kuttan G. Antiangiogenic activity of berberine is mediated through the downregulation of hypoxia-inducible factor-1, VEGF, and proinflammatory mediators. Drug Chem. Toxicol. 2012;35:57–70. doi: 10.3109/01480545.2011.589437. [DOI] [PubMed] [Google Scholar]
- 337.Kim S, et al. Berberine suppresses TPA-induced fibronectin expression through the inhibition of VEGF secretion in breast cancer cells. Cell Physiol. Biochem. 2013;32:1541–1550. doi: 10.1159/000356591. [DOI] [PubMed] [Google Scholar]
- 338.Ming J, et al. Effectiveness and safety of bifidobacteria and berberine in people with hyperglycemia: study protocol for a randomized controlled trial. Trials. 2018;19:72. doi: 10.1186/s13063-018-2438-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Cicero AF, Tartagni E. Antidiabetic properties of berberine: from cellular pharmacology to clinical effects. Hosp. Pract. (1995). 2012;40:56–63. doi: 10.3810/hp.2012.04.970. [DOI] [PubMed] [Google Scholar]
- 340.Lan J, et al. Meta-analysis of the effect and safety of berberine in the treatment of type 2 diabetes mellitus, hyperlipemia and hypertension. J. Ethnopharmacol. 2015;161:69–81. doi: 10.1016/j.jep.2014.09.049. [DOI] [PubMed] [Google Scholar]
- 341.Giovanelli A, Silva CL, Medeiros L, Vasconcellos MC. The molluscicidal activity of niclosamide (Bayluscide WP70(R)) on Melanoides tuberculata (Thiaridae), a snail associated with habitats of Biomphalaria glabrata (Planorbidae) Mem. Inst. Oswaldo Cruz. 2002;97:743–745. doi: 10.1590/s0074-02762002000500027. [DOI] [PubMed] [Google Scholar]
- 342.Pampori NA, Singh G, Srivastava VM. Cotugnia digonopora: carbohydrate metabolism and effect of anthelmintics on immature worms. J. Helminthol. 1984;58:39–47. doi: 10.1017/s0022149x00028042. [DOI] [PubMed] [Google Scholar]
- 343.Chen B, et al. Computational discovery of niclosamide ethanolamine, a repurposed drug candidate that reduces growth of hepatocellular carcinoma cells in vitro and in mice by inhibiting cell division cycle 37 signaling. Gastroenterology. 2017;152:2022–2036. doi: 10.1053/j.gastro.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Sack U, et al. Novel effect of antihelminthic niclosamide on S100A4-mediated metastatic progression in colon cancer. J. Natl Cancer Inst. 2011;103:1018–1036. doi: 10.1093/jnci/djr190. [DOI] [PubMed] [Google Scholar]
- 345.Lu L, et al. Activation of STAT3 and Bcl-2 and reduction of reactive oxygen species (ROS) promote radioresistance in breast cancer and overcome of radioresistance with niclosamide. Oncogene. 2018;37:5292–5304. doi: 10.1038/s41388-018-0340-y. [DOI] [PubMed] [Google Scholar]
- 346.King ML, et al. WNT7A/beta-catenin signaling induces FGF1 and influences sensitivity to niclosamide in ovarian cancer. Oncogene. 2015;34:3452–3462. doi: 10.1038/onc.2014.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Liao Z, et al. The anthelmintic drug niclosamide inhibits the proliferative activity of human osteosarcoma cells by targeting multiple signal pathways. Curr. Cancer Drug Targets. 2015;15:726–738. doi: 10.2174/1568009615666150629132157. [DOI] [PubMed] [Google Scholar]
- 348.Pan JX, Ding K, Wang CY. Niclosamide, an old antihelminthic agent, demonstrates antitumor activity by blocking multiple signaling pathways of cancer stem cells. Chin. J. Cancer. 2012;31:178–184. doi: 10.5732/cjc.011.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Dahlmann M, Kobelt D, Walther W, Mudduluru G, Stein U. S100A4 in cancer metastasisÿ: Wnt signaling-driven interventions for metastasis restriction. Cancers (Basel) 2016;8:59. doi: 10.3390/cancers8060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Stewart RL, et al. S100A4 drives non-small cell lung cancer invasion, associates with poor prognosis, and is effectively targeted by the FDA-approved anti-helminthic agent niclosamide. Oncotarget. 2016;7:34630–34642. doi: 10.18632/oncotarget.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Ye T, et al. The anthelmintic drug niclosamide induces apoptosis, impairs metastasis and reduces immunosuppressive cells in breast cancer model. PLoS ONE. 2014;9:e85887. doi: 10.1371/journal.pone.0085887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Liu J, et al. Niclosamide inhibits epithelial-mesenchymal transition and tumor growth in lapatinib-resistant human epidermal growth factor receptor 2-positive breast cancer. Int. J. Biochem. Cell Biol. 2016;71:12–23. doi: 10.1016/j.biocel.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 353.Guo C, et al. Targeting androgen receptor versus targeting androgens to suppress castration resistant prostate cancer. Cancer Lett. 2017;397:133–143. doi: 10.1016/j.canlet.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 354.Circu ML, et al. A novel high content imaging-based screen identifies the anti-helminthic niclosamide as an inhibitor of lysosome anterograde trafficking and prostate cancer cell invasion. PLoS ONE. 2016;11:e0146931. doi: 10.1371/journal.pone.0146931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wang LH, et al. The antihelminthic niclosamide inhibits cancer stemness, extracellular matrix remodeling, and metastasis through dysregulation of the nuclear beta-catenin/c-Myc axis in OSCC. Sci. Rep. 2018;8:12776. doi: 10.1038/s41598-018-30692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Chien MH, et al. Niclosamide, an oral antihelmintic drug, exhibits antimetastatic activity in hepatocellular carcinoma cells through downregulating twist-mediated CD10 expression. Environ. Toxicol. 2018;33:659–669. doi: 10.1002/tox.22551. [DOI] [PubMed] [Google Scholar]
- 357.Zhu Y, et al. Repurposing of the anti-helminthic drug niclosamide to treat melanoma and pulmonary metastasis via the STAT3 signaling pathway. Biochem. Pharmacol. 2019;169:113610. doi: 10.1016/j.bcp.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 358.Yu K, et al. Niclosamide induces apoptosis through mitochondrial intrinsic pathway and inhibits migration and invasion in human thyroid cancer in vitro. Biomed. Pharmacother. 2017;92:403–411. doi: 10.1016/j.biopha.2017.05.097. [DOI] [PubMed] [Google Scholar]
- 359.Chen H, et al. Discovery of O-alkylamino tethered niclosamide derivatives as potent and orally bioavailable anticancer agents. ACS Med. Chem. Lett. 2013;4:180–185. doi: 10.1021/ml3003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Ma R, et al. Injectable pegylated niclosamide (polyethylene glycol-modified niclosamide) for cancer therapy. J. Biomed. Mater. Res. A. 2020;108:30–38. doi: 10.1002/jbm.a.36788. [DOI] [PubMed] [Google Scholar]
- 361.Lin CK, et al. Preclinical evaluation of a nanoformulated antihelminthic, niclosamide, in ovarian cancer. Oncotarget. 2016;7:8993–9006. doi: 10.18632/oncotarget.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Tubbs A, Nussenzweig A, Endogenous DNA. Damage as a source of genomic instability in cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kim J, Bretz CL, Lee S. Epigenetic instability of imprinted genes in human cancers. Nucleic Acids Res. 2015;43:10689–10699. doi: 10.1093/nar/gkv867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 367.Pearl LH, et al. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 368.Helleday T, et al. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 369.Oxlund CS, et al. Low dose spironolactone reduces blood pressure in patients with resistant hypertension and type 2 diabetes mellitus: a double blind randomized clinical trial. J. Hypertens. 2013;31:2094–2102. doi: 10.1097/HJH.0b013e3283638b1a. [DOI] [PubMed] [Google Scholar]
- 370.Patibandla, S., Heaton, J. & Kyaw, H. in StatPearls (2020). [PubMed]
- 371.Manolis AA, Manolis TA, Melita H, Manolis AS. Spotlight on spironolactone oral suspension for the treatment of heart failure: focus on patient selection and perspectives. Vasc. Health Risk Manag. 2019;15:571–579. doi: 10.2147/VHRM.S210150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Mackenzie IS, et al. Spironolactone and risk of incident breast cancer in women older than 55 years: retrospective, matched cohort study. BMJ. 2012;345:e4447. doi: 10.1136/bmj.e4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Beckmann, K. et al. Spironolactone use is associated with lower prostate cancer risk: a population-wide case-control study. Prostate Cancer Prostatic Dis. 10.1038/s41391-020-0220-8 (2020). [DOI] [PubMed]
- 374.Alekseev S, et al. A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the chemosensitization of tumor cells to platinum. Chem. Biol. 2014;21:398–407. doi: 10.1016/j.chembiol.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 375.Ueda M, et al. Spironolactone-induced XPB degradation depends on CDK7 kinase and SCF(FBXL18) E3 ligase. Genes Cells. 2019;24:284–296. doi: 10.1111/gtc.12674. [DOI] [PubMed] [Google Scholar]
- 376.Zhang M, Du W, Acklin SM, Jin S, Xia F. SIRT2 protects peripheral neurons from cisplatin-induced injury by enhancing nucleotide excision repair. J. Clin. Invest. 2020;130:2953–2965. doi: 10.1172/JCI123159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Shahar OD, et al. A high-throughput chemical screen with FDA approved drugs reveals that the antihypertensive drug Spironolactone impairs cancer cell survival by inhibiting homology directed repair. Nucleic Acids Res. 2014;42:5689–5701. doi: 10.1093/nar/gku217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Gold A, et al. Spironolactone inhibits the growth of cancer stem cells by impairing DNA damage response. Oncogene. 2019;38:3103–3118. doi: 10.1038/s41388-018-0654-9. [DOI] [PubMed] [Google Scholar]
- 379.Guillotin D, et al. Drug-repositioning screens identify triamterene as a selective drug for the treatment of DNA mismatch repair deficient cells. Clin. Cancer Res. 2017;23:2880–2890. doi: 10.1158/1078-0432.CCR-16-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Keystone JS, Murdoch JK. Mebendazole. Ann. Intern. Med. 1979;91:582–586. doi: 10.7326/0003-4819-91-4-582. [DOI] [PubMed] [Google Scholar]
- 381.Sasaki J, et al. The anthelmintic drug mebendazole induces mitotic arrest and apoptosis by depolymerizing tubulin in non-small cell lung cancer cells. Mol. Cancer Ther. 2002;1:1201–1209. [PubMed] [Google Scholar]
- 382.Bai RY, et al. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neuro Oncol. 2011;13:974–982. doi: 10.1093/neuonc/nor077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Pinto LC, et al. The anthelmintic drug mebendazole inhibits growth, migration and invasion in gastric cancer cell model. Toxicol. Vitr. 2015;29:2038–2044. doi: 10.1016/j.tiv.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 384.Markowitz D, Ha G, Ruggieri R, Symons M. Microtubule-targeting agents can sensitize cancer cells to ionizing radiation by an interphase-based mechanism. Onco Targets Ther. 2017;10:5633–5642. doi: 10.2147/OTT.S143096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Zhang L, et al. Mebendazole potentiates radiation therapy in triple-negative breast cancer. Int J. Radiat. Oncol. Biol. Phys. 2019;103:195–207. doi: 10.1016/j.ijrobp.2018.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Larsen AR, et al. Repurposing the antihelmintic mebendazole as a hedgehog inhibitor. Mol. Cancer Ther. 2015;14:3–13. doi: 10.1158/1535-7163.MCT-14-0755-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 388.Williamson T, et al. Mebendazole inhibits tumor growth and prevents lung metastasis in models of advanced thyroid cancer. Endocr. Relat. Cancer. 2020;27:123–136. doi: 10.1530/ERC-19-0341. [DOI] [PubMed] [Google Scholar]
- 389.Zhang F, et al. Anthelmintic mebendazole enhances cisplatin's effect on suppressing cell proliferation and promotes differentiation of head and neck squamous cell carcinoma (HNSCC) Oncotarget. 2017;8:12968–12982. doi: 10.18632/oncotarget.14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Abdullah I, et al. Benzimidazole derivatives as potential dual inhibitors for PARP-1 and DHODH. Bioorg. Med. Chem. 2015;23:4669–4680. doi: 10.1016/j.bmc.2015.05.051. [DOI] [PubMed] [Google Scholar]
- 391.Dobrosotskaya IY, et al. Mebendazole monotherapy and long-term disease control in metastatic adrenocortical carcinoma. Endocr. Pract. 2011;17:e59–e62. doi: 10.4158/EP10390.CR. [DOI] [PubMed] [Google Scholar]
- 392.Nygren P, Larsson R. Drug repositioning from bench to bedside: tumour remission by the antihelmintic drug mebendazole in refractory metastatic colon cancer. Acta Oncol. 2014;53:427–428. doi: 10.3109/0284186X.2013.844359. [DOI] [PubMed] [Google Scholar]
- 393.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15:e493–e503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 394.Bondar T, Medzhitov R. The origins of tumor-promoting inflammation. Cancer Cell. 2013;24:143–144. doi: 10.1016/j.ccr.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.El-Kenawi A, Ruffell B. Inflammation, ROS, and mutagenesis. Cancer Cell. 2017;32:727–729. doi: 10.1016/j.ccell.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 397.Vane JR. The mode of action of aspirin and similar compounds. J. Allergy Clin. Immunol. 1976;58:691–712. doi: 10.1016/0091-6749(76)90181-0. [DOI] [PubMed] [Google Scholar]
- 398.Gaziano JM, et al. Use of aspirin to reduce risk of initial vascular events in patients at moderate risk of cardiovascular disease (ARRIVE): a randomised, double-blind, placebo-controlled trial. Lancet. 2018;392:1036–1046. doi: 10.1016/S0140-6736(18)31924-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Amarenco P, et al. Efficacy and safety of ticagrelor versus aspirin in acute stroke or transient ischaemic attack of atherosclerotic origin: a subgroup analysis of SOCRATES, a randomised, double-blind, controlled trial. Lancet Neurol. 2017;16:301–310. doi: 10.1016/S1474-4422(17)30038-8. [DOI] [PubMed] [Google Scholar]
- 400.Gasic GJ, et al. Platelet-tumor-cell interactions in mice. The role of platelets in the spread of malignant disease. Int J. Cancer. 1973;11:704–718. doi: 10.1002/ijc.2910110322. [DOI] [PubMed] [Google Scholar]
- 401.Kalgutkar AS, et al. Aspirin-like molecules that covalently inactivate cyclooxygenase-2. Science. 1998;280:1268–1270. doi: 10.1126/science.280.5367.1268. [DOI] [PubMed] [Google Scholar]
- 402.Wang D, Dubois RN. Prostaglandins and cancer. Gut. 2006;55:115–122. doi: 10.1136/gut.2004.047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Kumar D, et al. Aspirin suppresses PGE2 and activates AMP kinase to inhibit melanoma cell motility, pigmentation, and selective tumor growth in vivo. Cancer Prev. Res (Philos.). 2018;11:629–642. doi: 10.1158/1940-6207.CAPR-18-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Kashiwagi E, et al. Prostaglandin receptor EP3 mediates growth inhibitory effect of aspirin through androgen receptor and contributes to castration resistance in prostate cancer cells. Endocr. Relat. Cancer. 2013;20:431–441. doi: 10.1530/ERC-12-0344. [DOI] [PubMed] [Google Scholar]
- 405.Barnard ME, et al. Urinary PGE-M levels and risk of ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2019;28:1845–1852. doi: 10.1158/1055-9965.EPI-19-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Li M, et al. Aspirin induction of apoptosis in esophageal cancer: a potential for chemoprevention. Cancer Epidemiol. Biomark. Prev. 2000;9:545–549. [PubMed] [Google Scholar]
- 407.Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat. Rev. Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 408.Zhang Z, Huang L, Zhao W, Rigas B. Annexin 1 induced by anti-inflammatory drugs binds to NF-kappaB and inhibits its activation: anticancer effects in vitro and in vivo. Cancer Res. 2010;70:2379–2388. doi: 10.1158/0008-5472.CAN-09-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Huo X, et al. Aspirin prevents NF-kappaB activation and CDX2 expression stimulated by acid and bile salts in oesophageal squamous cells of patients with Barrett's oesophagus. Gut. 2018;67:606–615. doi: 10.1136/gutjnl-2016-313584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.O'Hara A, et al. p300-mediated acetylation of COMMD1 regulates its stability, and the ubiquitylation and nucleolar translocation of the RelA NF-kappaB subunit. J. Cell Sci. 2014;127:3659–3665. doi: 10.1242/jcs.149328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Hsieh CC, Huang YS. Aspirin breaks the crosstalk between 3T3-L1 adipocytes and 4T1 breast cancer cells by regulating cytokine production. PLoS ONE. 2016;11:e0147161. doi: 10.1371/journal.pone.0147161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Hsieh CC, Wang CH. Aspirin disrupts the crosstalk of angiogenic and inflammatory cytokines between 4T1 breast cancer cells and macrophages. Mediators Inflamm. 2018;2018:6380643. doi: 10.1155/2018/6380643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Gilligan MM, et al. Aspirin-triggered proresolving mediators stimulate resolution in cancer. Proc. Natl Acad. Sci. USA. 2019;116:6292–6297. doi: 10.1073/pnas.1804000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Kraus S, Sion D, Arber N. Can we select patients for colorectal cancer prevention with aspirin? Curr. Pharm. Des. 2015;21:5127–5134. doi: 10.2174/1381612821666150915111000. [DOI] [PubMed] [Google Scholar]
- 415.Stegeman I, et al. Aspirin for primary prevention of cardiovascular disease and cancer. a benefit and harm analysis. PLoS ONE. 2015;10:e0127194. doi: 10.1371/journal.pone.0127194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Rothwell PM, et al. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377:31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- 417.Drew DA, Cao Y, Chan AT. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat. Rev. Cancer. 2016;16:173–186. doi: 10.1038/nrc.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Liao X, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N. Engl. J. Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Li P, et al. Aspirin use after diagnosis but not prediagnosis improves established colorectal cancer survival: a meta-analysis. Gut. 2015;64:1419–1425. doi: 10.1136/gutjnl-2014-308260. [DOI] [PubMed] [Google Scholar]
- 420.Tuzun F, et al. Multicenter, randomized, double-blinded, placebo-controlled trial of thiocolchicoside in acute low back pain. Jt. Bone Spine. 2003;70:356–361. doi: 10.1016/s1297-319x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 421.Reuter S, et al. Thiocolchicoside exhibits anticancer effects through downregulation of NF-kappaB pathway and its regulated gene products linked to inflammation and cancer. Cancer Prev. Res. (Philos.). 2010;3:1462–1472. doi: 10.1158/1940-6207.CAPR-10-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Reuter S, Gupta SC, Phromnoi K, Aggarwal BB. Thiocolchicoside suppresses osteoclastogenesis induced by RANKL and cancer cells through inhibition of inflammatory pathways: a new use for an old drug. Br. J. Pharmacol. 2012;165:2127–2139. doi: 10.1111/j.1476-5381.2011.01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Micheau O, Dufour F, Walczak H. Thiocolchicoside a semi-synthetic derivative of the Glory Lily: a new weapon to fight metastatic bone resorption? Br. J. Pharmacol. 2012;165:2124–2126. doi: 10.1111/j.1476-5381.2011.01792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Sproviero E, et al. Efficacy and safety of a fixed combination of intramuscular diclofenac 75 mg + thiocolchicoside 4 mg in the treatment of acute low back pain: a phase III, randomized, double blind, controlled trial. Eur. J. Phys. Rehabil. Med. 2018;54:654–662. doi: 10.23736/S1973-9087.17.04923-1. [DOI] [PubMed] [Google Scholar]
- 425.Rao R, et al. Clinical comparative study: efficacy and tolerability of tolperisone and thiocolchicoside in acute low back pain and spinal muscle spasticity. Asian Spine J. 2012;6:115–122. doi: 10.4184/asj.2012.6.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Russo M, et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473–1480. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- 427.Wicki A, et al. Acquired resistance to clinical cancer therapy: a twist in physiological signaling. Physiol. Rev. 2016;96:805–829. doi: 10.1152/physrev.00024.2015. [DOI] [PubMed] [Google Scholar]
- 428.Sleire L, et al. Drug repurposing in cancer. Pharm. Res. 2017;124:74–91. doi: 10.1016/j.phrs.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 429.Saini N, Yang X. Metformin as an anticancer agent: actions and mechanisms targeting cancer stem cells. Acta Biochim. Biophys. Sin. 2018;50:133–143. doi: 10.1093/abbs/gmx106. [DOI] [PubMed] [Google Scholar]
- 430.Jiao Y, Hannafon BN, Ding WQ. Disulfiram's anticancer activity: evidence and mechanisms. Anticancer Agents Med. Chem. 2016;16:1378–1384. doi: 10.2174/1871520615666160504095040. [DOI] [PubMed] [Google Scholar]
- 431.Zhou L, et al. Brefeldin A inhibits colorectal cancer growth by triggering Bip/Akt-regulated autophagy. FASEB J. 2019;33:5520–5534. doi: 10.1096/fj.201801983R. [DOI] [PubMed] [Google Scholar]
- 432.Chen Y, et al. Ketoconazole exacerbates mitophagy to induce apoptosis by downregulating cyclooxygenase-2 in hepatocellular carcinoma. J. Hepatol. 2019;70:66–77. doi: 10.1016/j.jhep.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 433.Dou Q, et al. Ivermectin induces cytostatic autophagy by blocking the PAK1/Akt axis in breast cancer. Cancer Res. 2016;76:4457–4469. doi: 10.1158/0008-5472.CAN-15-2887. [DOI] [PubMed] [Google Scholar]
- 434.Bray M, Rayner C, Noel F, Jans D, Wagstaff K. Ivermectin and COVID-19: a report in Antiviral Research, widespread interest, an FDA warning, two letters to the editor and the authors' responses. Antiviral Res. 2020;178:104805. doi: 10.1016/j.antiviral.2020.104805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Caly L, et al. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.El-Hachem N, et al. Integrative cancer pharmacogenomics to infer large-scale drug taxonomy. Cancer Res. 2017;77:3057–3069. doi: 10.1158/0008-5472.CAN-17-0096. [DOI] [PubMed] [Google Scholar]
- 437.Luo Y, et al. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 2017;8:573. doi: 10.1038/s41467-017-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Fernandez-Torras A, Duran-Frigola M, Aloy P. Encircling the regions of the pharmacogenomic landscape that determine drug response. Genome Med. 2019;11:17. doi: 10.1186/s13073-019-0626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Mottini, C., Napolitano, F., Li, Z., Gao, X. & Cardone, L. Computer-aided drug repurposing for cancer therapy: approaches and opportunities to challenge anticancer targets. Semin. Cancer Biol.10.1016/j.semcancer.2019.09.023 (2019). [DOI] [PubMed]
- 440.Palve, V., Liao, Y., Remsing Rix, L. L. & Rix, U. Turning liabilities into opportunities: off-target based drug repurposing in cancer. Semin. Cancer Biol.10.1016/j.semcancer.2020.02.003 (2020). [DOI] [PMC free article] [PubMed]
- 441.Guney E, Menche J, Vidal M, Barabasi AL. Network-based in silico drug efficacy screening. Nat. Commun. 2016;7:10331. doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Li YY, Jones SJ. Drug repositioning for personalized medicine. Genome Med. 2012;4:27. doi: 10.1186/gm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Issa, N. T., Stathias, V., Schurer, S. & Dakshanamurthy, S. Machine and deep learning approaches for cancer drug repurposing. Semin. Cancer Biol. 10.1016/j.semcancer.2019.12.011 (2020). [DOI] [PMC free article] [PubMed]
- 444.Nowak-Sliwinska P, Scapozza L, Ruiz IAA. Drug repurposing in oncology: compounds, pathways, phenotypes and computational approaches for colorectal cancer. Biochim. Biophys. Acta Rev. Cancer. 2019;1871:434–454. doi: 10.1016/j.bbcan.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Jones LH, Bunnage ME. Applications of chemogenomic library screening in drug discovery. Nat. Rev. Drug Discov. 2017;16:285–296. doi: 10.1038/nrd.2016.244. [DOI] [PubMed] [Google Scholar]
- 446.Kuhlman B, Bradley P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019;20:681–697. doi: 10.1038/s41580-019-0163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Niu B, et al. Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet. 2016;48:827–837. doi: 10.1038/ng.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Knapp S. New opportunities for kinase drug repurposing and target discovery. Br. J. Cancer. 2018;118:936–937. doi: 10.1038/s41416-018-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Ma DL, Chan DS, Leung CH. Drug repositioning by structure-based virtual screening. Chem. Soc. Rev. 2013;42:2130–2141. doi: 10.1039/c2cs35357a. [DOI] [PubMed] [Google Scholar]
- 450.Gorgulla C, et al. An open-source drug discovery platform enables ultra-large virtual screens. Nature. 2020;580:663–668. doi: 10.1038/s41586-020-2117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Honarparvar B, et al. Integrated approach to structure-based enzymatic drug design: molecular modeling, spectroscopy, and experimental bioactivity. Chem. Rev. 2014;114:493–537. doi: 10.1021/cr300314q. [DOI] [PubMed] [Google Scholar]
- 452.Nero TL, et al. Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 453.Salentin S, et al. From malaria to cancer: computational drug repositioning of amodiaquine using PLIP interaction patterns. Sci. Rep. 2017;7:11401. doi: 10.1038/s41598-017-11924-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Congiatu C, Brancale A, McGuigan C. Molecular modelling studies on the binding of some protides to the putative human phosphoramidase Hint1. Nucleosides Nucleotides Nucleic Acids. 2007;26:1121–1124. doi: 10.1080/15257770701521656. [DOI] [PubMed] [Google Scholar]
- 455.Heinrich JC, et al. New HSP27 inhibitors efficiently suppress drug resistance development in cancer cells. Oncotarget. 2016;7:68156–68169. doi: 10.18632/oncotarget.11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Heinrich JC, et al. RP101 (brivudine) binds to heat shock protein HSP27 (HSPB1) and enhances survival in animals and pancreatic cancer patients. J. Cancer Res. Clin. Oncol. 2011;137:1349–1361. doi: 10.1007/s00432-011-1005-1. [DOI] [PubMed] [Google Scholar]
- 457.Stular T, et al. Discovery of mycobacterium tuberculosis InhA inhibitors by binding sites comparison and ligands prediction. J. Med. Chem. 2016;59:11069–11078. doi: 10.1021/acs.jmedchem.6b01277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Salentin S, et al. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Lim H, et al. Rational discovery of dual-indication multi-target PDE/Kinase inhibitor for precision anticancer therapy using structural systems pharmacology. PLoS Comput Biol. 2019;15:e1006619. doi: 10.1371/journal.pcbi.1006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Fernandez-Banet J, et al. OASIS: web-based platform for exploring cancer multi-omics data. Nat. Methods. 2016;13:9–10. doi: 10.1038/nmeth.3692. [DOI] [PubMed] [Google Scholar]
- 461.Chen B, et al. Harnessing big ‘omics' data and AI for drug discovery in hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2020;17:238–251. doi: 10.1038/s41575-019-0240-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat. Rev. Cancer. 2017;17:692–704. doi: 10.1038/nrc.2017.82. [DOI] [PubMed] [Google Scholar]
- 463.Kim J, et al. Systems pharmacology-based approach of connecting disease genes in genome-wide association studies with traditional Chinese medicine. Int. J. Genom. 2018;2018:7697356. doi: 10.1155/2018/7697356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Zhang J, et al. Use of genome-wide association studies for cancer research and drug repositioning. PLoS ONE. 2015;10:e0116477. doi: 10.1371/journal.pone.0116477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Mei, S. Zhang, K. A Multi-label learning framework for drug repurposing. Pharmaceutics11 (2019). [DOI] [PMC free article] [PubMed]
- 466.Singh VK, et al. Drug repurposing for chronic myeloid leukemia: in silico and in vitro investigation of DrugBank database for allosteric Bcr-Abl inhibitors. J. Biomol. Struct. Dyn. 2017;35:1833–1848. doi: 10.1080/07391102.2016.1196462. [DOI] [PubMed] [Google Scholar]
- 467.You J, McLeod RD, Hu P. Predicting drug-target interaction network using deep learning model. Comput. Biol. Chem. 2019;80:90–101. doi: 10.1016/j.compbiolchem.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 468.Wei WQ, Mosley JD, Bastarache L, Denny JC. Validation and enhancement of a computable medication indication resource (MEDI) using a large practice-based dataset. AMIA Annu. Symp. Proc. 2013;2013:1448–1456. [PMC free article] [PubMed] [Google Scholar]
- 469.Bejan CA, Wei WQ, Denny JC. Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text. J. Am. Med Inform. 2015;22:e162–e176. doi: 10.1136/amiajnl-2014-002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Bielefeldt K. Ischemic colitis as a complication of medication use: an analysis of the federal adverse event reporting system. Dig. Dis. Sci. 2016;61:2655–2665. doi: 10.1007/s10620-016-4162-x. [DOI] [PubMed] [Google Scholar]
- 471.Camilleri M. Implications of pharmacogenomics to the management of IBS. Clin. Gastroenterol. Hepatol. 2019;17:584–594. doi: 10.1016/j.cgh.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 472.Sertznig P, et al. Cross-talk between vitamin D receptor (VDR)- and peroxisome proliferator-activated receptor (PPAR)-signaling in melanoma cells. Anticancer Res. 2009;29:3647–3658. [PubMed] [Google Scholar]
- 473.Xu C, et al. Accurate drug repositioning through non-tissue-specific core signatures from cancer transcriptomes. Cell Rep. 2018;25:523–535. doi: 10.1016/j.celrep.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 474.Szalai B, et al. Signatures of cell death and proliferation in perturbation transcriptomics data-from confounding factor to effective prediction. Nucleic Acids Res. 2019;47:10010–10026. doi: 10.1093/nar/gkz805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Booth CM, Karim S, Mackillop WJ. Real-world data: towards achieving the achievable in cancer care. Nat. Rev. Clin. Oncol. 2019;16:312–325. doi: 10.1038/s41571-019-0167-7. [DOI] [PubMed] [Google Scholar]
- 476.Dickson D, et al. Snapshot: trial types in precision medicine. Cell. 2020;181:208–208. doi: 10.1016/j.cell.2020.02.032. [DOI] [PubMed] [Google Scholar]
- 477.Eichler HG, et al. Data rich, information poor: can we use electronic health records to create a learning healthcare system for pharmaceuticals? Clin. Pharm. Ther. 2019;105:912–922. doi: 10.1002/cpt.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Dai HJ, Syed-Abdul S, Chen CW, Wu CC. Recognition and evaluation of clinical section headings in clinical documents using token-based formulation with conditional random fields. Biomed. Res. Int. 2015;2015:873012. doi: 10.1155/2015/873012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Luhn P, et al. Validation of diagnosis codes to identify side of colon in an electronic health record registry. BMC Med. Res. Methodol. 2019;19:177. doi: 10.1186/s12874-019-0824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Miriovsky BJ, Shulman LN, Abernethy AP. Importance of health information technology, electronic health records, and continuously aggregating data to comparative effectiveness research and learning health care. J. Clin. Oncol. 2012;30:4243–4248. doi: 10.1200/JCO.2012.42.8011. [DOI] [PubMed] [Google Scholar]
- 481.Khozin, S., Blumenthal, G. M. & Pazdur, R. Real-world data for clinical evidence generation in oncology. J. Natl Cancer Inst. 109, 10.1093/jnci/djx187 (2017). [DOI] [PubMed]
- 482.Low YS, et al. Synergistic drug combinations from electronic health records and gene expression. J. Am. Med. Inform. Assoc. 2017;24:565–576. doi: 10.1093/jamia/ocw161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Ventz S, et al. Design and evaluation of an external control arm using prior clinical trials and real-world data. Clin. Cancer Res. 2019;25:4993–5001. doi: 10.1158/1078-0432.CCR-19-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Marchenko O, et al. Sources of safety data and statistical strategies for design and analysis: real world insights. Ther. Innov. Regul. Sci. 2018;52:170–186. doi: 10.1177/2168479017739270. [DOI] [PubMed] [Google Scholar]
- 485.Zheng S, et al. Identifying the characteristics of patients with cervical degenerative disease for surgical treatment from 17-year real-world data: retrospective study. JMIR Med. Inform. 2020;8:e16076. doi: 10.2196/16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Gray E, et al. Chemotherapy effectiveness in trial-underrepresented groups with early breast cancer: a retrospective cohort study. PLoS Med. 2019;16:e1003006. doi: 10.1371/journal.pmed.1003006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Chaiteerakij R, et al. Metformin use and survival of patients with pancreatic cancer: a cautionary lesson. J. Clin. Oncol. 2016;34:1898–1904. doi: 10.1200/JCO.2015.63.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Sadeghi N, et al. Metformin use is associated with better survival of diabetic patients with pancreatic cancer. Clin. Cancer Res. 2012;18:2905–2912. doi: 10.1158/1078-0432.CCR-11-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Xu H, et al. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J. Am. Med. Inform. Assoc. 2015;22:179–191. doi: 10.1136/amiajnl-2014-002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Happold C, et al. Does valproic acid or levetiracetam improve survival in glioblastoma? A pooled analysis of prospective clinical trials in newly diagnosed glioblastoma. J. Clin. Oncol. 2016;34:731–739. doi: 10.1200/JCO.2015.63.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Blum W, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J. Clin. Oncol. 2007;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 492.Lubbert M, et al. Valproate and retinoic acid in combination with decitabine in elderly nonfit patients with acute myeloid leukemia: results of a multicenter, randomized, 2 x 2, phase II trial. J. Clin. Oncol. 2020;38:257–270. doi: 10.1200/JCO.19.01053. [DOI] [PubMed] [Google Scholar]
- 493.Perez L, et al. Application of liquid biopsies in metastatic gastrointestinal cancer to identify candidate therapeutic targets. Ann. Oncol. 2018;29(Suppl 6):vi1. [Google Scholar]
- 494.Shantikumar S, Satheeshkumar N, Srinivas R. Pharmacokinetic and protein binding profile of peptidomimetic DPP-4 inhibitor—teneligliptin in rats using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015;1002:194–200. doi: 10.1016/j.jchromb.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 495.Taguchi A, et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell. 2011;20:289–299. doi: 10.1016/j.ccr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Savitski MM, et al. Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell. 2018;173:260–274. doi: 10.1016/j.cell.2018.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Schwarzl T, Higgins DG, Kolch W, Duffy DJ. Measuring transcription rate changes via time-course 4-thiouridine pulse-labelling improves transcriptional target identification. J. Mol. Biol. 2015;427:3368–3374. doi: 10.1016/j.jmb.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 498.Boros LG, Brackett DJ, Harrigan GG. Metabolic biomarker and kinase drug target discovery in cancer using stable isotope-based dynamic metabolic profiling (SIDMAP) Curr. Cancer Drug Targets. 2003;3:445–453. doi: 10.2174/1568009033481769. [DOI] [PubMed] [Google Scholar]
- 499.Brehmer D, et al. Cellular targets of gefitinib. Cancer Res. 2005;65:379–382. [PubMed] [Google Scholar]
- 500.Li J, et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods. 2020;17:399–404. doi: 10.1038/s41592-020-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 502.Mateus A, et al. Thermal proteome profiling for interrogating protein interactions. Mol. Syst. Biol. 2020;16:e9232. doi: 10.15252/msb.20199232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Franken H, et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 504.Perrin J, et al. Identifying drug targets in tissues and whole blood with thermal-shift profiling. Nat. Biotechnol. 2020;38:303–308. doi: 10.1038/s41587-019-0388-4. [DOI] [PubMed] [Google Scholar]
- 505.Cong F, Cheung AK, Huang SM. Chemical genetics-based target identification in drug discovery. Annu Rev. Pharm. Toxicol. 2012;52:57–78. doi: 10.1146/annurev-pharmtox-010611-134639. [DOI] [PubMed] [Google Scholar]
- 506.Wong LH, et al. Reverse chemical genetics: comprehensive fitness profiling reveals the spectrum of drug target interactions. PLoS Genet. 2016;12:e1006275. doi: 10.1371/journal.pgen.1006275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 508.Sala C, et al. Simple model for testing drugs against nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2010;54:4150–4158. doi: 10.1128/AAC.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Iljin K, et al. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin. Cancer Res. 2009;15:6070–6078. doi: 10.1158/1078-0432.CCR-09-1035. [DOI] [PubMed] [Google Scholar]
- 510.Corsello SM, et al. Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer. 2020;1:235–248. doi: 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Beijersbergen RL. Old drugs with new tricks. Nat. Cancer. 2020;1:153–155. doi: 10.1038/s43018-020-0024-8. [DOI] [PubMed] [Google Scholar]
- 512.Yadav AK, Srikrishna S, Gupta SC. Cancer drug development using drosophila as an in vivo tool: from bedside to bench and back. Trends Pharm. Sci. 2016;37:789–806. doi: 10.1016/j.tips.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 513.Clements WK, Traver D. Fish pharming: zebrafish antileukemia screening. Blood. 2012;119:5614–5615. doi: 10.1182/blood-2012-04-425249. [DOI] [PubMed] [Google Scholar]
- 514.Ridges S, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119:5621–5631. doi: 10.1182/blood-2011-12-398818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Baxendale S, van Eeden F, Wilkinson R. The power of zebrafish in personalised medicine. Adv. Exp. Med. Biol. 2017;1007:179–197. doi: 10.1007/978-3-319-60733-7_10. [DOI] [PubMed] [Google Scholar]
- 516.Gonzalez-Fierro, A. & Duenas-Gonzalez, A. Drug repurposing for cancer therapy, easier said than done. Semin. Cancer Biol.10.1016/j.semcancer.2019.12.012 (2019). [DOI] [PubMed]
- 517.Breckenridge A, Jacob R. Overcoming the legal and regulatory barriers to drug repurposing. Nat. Rev. Drug Discov. 2019;18:1–2. doi: 10.1038/nrd.2018.92. [DOI] [PubMed] [Google Scholar]
- 518.Pantziarka P, et al. The repurposing drugs in oncology (ReDO) project. Ecancermedicalscience. 2014;8:442. doi: 10.3332/ecancer.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Stewart AK, et al. Melphalan, prednisone, and thalidomide vs melphalan, prednisone, and lenalidomide (ECOG E1A06) in untreated multiple myeloma. Blood. 2015;126:1294–1301. doi: 10.1182/blood-2014-12-613927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Zweegman S, et al. Melphalan, prednisone, and lenalidomide versus melphalan, prednisone, and thalidomide in untreated multiple myeloma. Blood. 2016;127:1109–1116. doi: 10.1182/blood-2015-11-679415. [DOI] [PubMed] [Google Scholar]
- 521.Sullivan, T. A tough road: cost to develop one new drug is $2.6 billion; approval rate for drugs entering clinical development is less than 12%. https://www.policymed.com/2014/12/a-tough-road-cost-to-develop-one-new-drug-is-26-billion-approval-rate-for-drugs-entering-clinical-de.html (2019).
- 522.Raju TN. The Nobel chronicles. 1988: James Whyte Black, (b 1924), Gertrude Elion (1918–99), and George H Hitchings (1905–98) Lancet. 2000;355:1022. doi: 10.1016/s0140-6736(05)74775-9. [DOI] [PubMed] [Google Scholar]