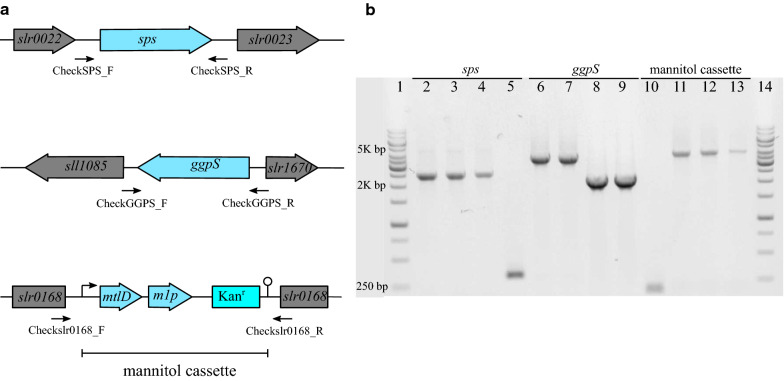
Fig. 2.
Identification and confirmation genotype by PCR in the Synechocystis genome. a Map of primer-binding sites. Primers CheckSPS_F and CheckSPS_R were used for segregation analysis of the sps locus (PCR products 300 bp in mutant and 2400 bp in WT). Primers CheckGGPS_F and CheckGGPS_R were used to check the ggpS locus (PCR products 2100 bp in mutant and 4800 bp in WT). Primers Checkslr0168_F and Checkslr0168_R were used to check the slr0168 locus (PCR products 5200 bp in mutant and 200 bp in WT). b PCR analyses for genotyping of mutants. The chromosomal DNA is from WT (lanes 2, 6 and 10), WT_M (lanes 3,7 and 11), ΔGGPS_M (lanes 4,8 and 12) and ΔCS_M (lanes 5,9 and 13) of Synechocystis as a template and primers CheckSPS_F and CheckSPS_R specific for selected genes sps (lanes 2 to 5), primers CheckGGPS_F and CheckGGPS_R specific for selected genes ggpS (lanes 6 to 9), and primers Checkslr0168_F and Checkslr0168_R specific for selected genes mannitol cassette (lanes 10 to 13) to verify the genotype in the chromosomal DNA of the mutants. Lanes 1 and 14 ladder (1 kb from Fisher Scientific Company). Kanr, indicates kanamycin resistance cassette