Abstract
While severe coronavirus infections, including Middle East respiratory syndrome coronavirus (MERS-CoV), cause lung injury with high mortality rates, protective treatment strategies are not approved for clinical use.
We elucidated the molecular mechanisms by which the cyclophilin inhibitors cyclosporin A (CsA) and alisporivir (ALV) restrict MERS-CoV to validate their suitability as readily available therapy in MERS-CoV infection.
Calu-3 cells and primary human alveolar epithelial cells (hAECs) were infected with MERS-CoV and treated with CsA or ALV or inhibitors targeting cyclophilin inhibitor-regulated molecules including calcineurin, nuclear factor of activated T-cells (NFATs) or mitogen-activated protein kinases. Novel CsA-induced pathways were identified by RNA sequencing and manipulated by gene knockdown or neutralising antibodies. Viral replication was quantified by quantitative real-time PCR and 50% tissue culture infective dose. Data were validated in a murine MERS-CoV infection model.
Both CsA and ALV reduced MERS-CoV titres and viral RNA replication in Calu-3 cells and hAECs, improving epithelial integrity. While neither calcineurin nor NFAT inhibition reduced MERS-CoV propagation, blockade of c-Jun N-terminal kinase diminished infectious viral particle release but not RNA accumulation. Importantly, CsA induced interferon regulatory factor 1 (IRF1), a pronounced type III interferon (IFNλ) response and expression of antiviral genes. Downregulation of IRF1 or IFNλ increased MERS-CoV propagation in the presence of CsA. Importantly, oral application of CsA reduced MERS-CoV replication in vivo, correlating with elevated lung IFNλ levels and improved outcome.
We provide evidence that cyclophilin inhibitors efficiently decrease MERS-CoV replication in vitro and in vivo via upregulation of inflammatory antiviral cell responses, in particular IFNλ. CsA might therefore represent a promising candidate for treating MERS-CoV infection.
Short abstract
The cyclophilin inhibitors cyclosporin A and alisporivir activate host innate immunity by induction of interferon-λ via activation of IRF1 in human lung epithelial cells and in vivo, resulting in a significant inhibition of MERS-CoV https://bit.ly/37gzIBH
Introduction
Middle East respiratory syndrome coronavirus (MERS-CoV) emerged in 2012 in Saudi Arabia [1] and led to recurring human infections with more than 2500 laboratory-confirmed cases and high case fatality rates of about 35% [2]. In ex vivo infection of human lung tissue, MERS-CoV targets bronchial and alveolar epithelial cells (AECs) and leads to a detachment and apoptosis of AECs [3]. Recent reports analysing autopsy material from deceased MERS-CoV-infected patients showed MERS-CoV antigen in AECs and epithelial multinucleated syncytial cell conglomerates in vivo [4, 5]. Accordingly, severe human infection presents as pneumonia with progression to acute respiratory distress syndrome [4, 5].
To date, no vaccine or specific treatment for MERS-CoV, or the pandemic novel severe acute respiratory syndrome CoV 2 (SARS-CoV-2), has been approved and therapy relies on supportive measures only [2, 6]. While in vitro studies and experiments in non-human primates demonstrated benefits of a combination of type I interferon (IFN) and antiviral compounds, including ribavirin, against MERS-CoV [7–9], results from retrospective patient cohorts applying similar treatment regimens remain controversial [10–12]. Cyclosporin A (CsA) has been found to inhibit several human-pathogenic CoV in cell lines originating from kidney or liver epithelia [13–16]. However, the molecular mechanisms by which CsA affects CoV, including MERS-CoV, particularly in its primary target cells, the pulmonary epithelium, remain elusive. Moreover, preclinical studies addressing the effect of CsA or related compounds on MERS-CoV replication in vivo have been lacking to date.
CsA is known to block the peptidyl-prolyl cis-trans isomerase (PPI) activity of cyclophilins that is involved in diverse cellular processes (e.g. protein folding [17]). Additionally, CsA forms a ternary complex with cyclophilin A (CypA) and calcineurin (CnA) that blocks the CnA-dependent activation of nuclear factor of activated T-cells (NFATs), a process that accounts for the immunosuppressive effect of CsA [18]. CsA has also been shown to inhibit the mitogen-activated protein kinases (MAPKs) c-Jun N-terminal kinase (JNK) and p38 [19, 20].
Here, we aimed to elucidate the distinct signalling pathways by which CsA affects MERS-CoV in clinically relevant models such as primary human AECs (hAECs) and a murine MERS-CoV infection model [21, 22]. We demonstrate that CsA blocks MERS-CoV infectious particle egress, which is dependent on JNK. Moreover, we for the first time provide evidence that CsA triggers the activation of an antiviral defence state in lung epithelial cells. We show that CsA is a potent inducer of interferon regulatory factor 1 (IRF1), type III IFN (IFNλ) and multiple interferon-stimulated genes (ISGs). Additionally, we demonstrate that oral application of CsA induces a robust IFNλ response in vivo and, importantly, significantly reduces MERS-CoV replication and improves disease progression in infected mice.
Methods
MERS-CoV infection
Experiments with MERS-CoV were performed under biosafety level 4 conditions at the Institute of Virology, Philipps University of Marburg, Germany. hAECs were isolated and cultured as previously described [23]. Human lung tissue was obtained from patients who underwent lobectomy after informed written consent (Depts of Pathology and Surgery, University of Giessen, Germany, approved by the University of Giessen Ethics Committee; Az.58/15). Calu-3 cells or hAECs were infected at a multiplicity of infection of 0.1 diluted in DMEM/F12 without fetal calf serum (FCS) at 37°C for 1 h. Cells were washed with DMEM/F12 with 10% FCS and supplemented with stimulatory/inhibitory reagents as indicated. 24 h post infection, cells were processed for quantitative PCR (Maxima-SYBR/ROX qPCR-Mastermix, Thermo Fisher Scientific, Waltham, MA, USA) and the supernatant was harvested for virus titration as described previously [24].
In vivo transduction and infection
All animal experiments were performed in accordance with the German animal protection laws and were authorised by the regional authorities (G73/2017). C57BL/6 mice were purchased from Charles River Laboratories (Wilmington, MA, USA) and housed under pathogen-free conditions. Mice underwent intratracheal inoculation with Adenovirus-hDPP4-mCherry (cloned at ViraQuest Inc., North Liberty, IA, USA) as described [21, 25]. 5 days post transduction, mice were infected via the intranasal route with a 50% tissue culture infective dose (TCID50) at 1.5×105 TCID50·mL−1 of MERS-CoV EMC/2012. 50 mg·kg−1·day−1 CsA diluted in DMSO or DMSO alone were mixed with a nut/chocolate cream, and offered to the mice for voluntary uptake. Uptake was controlled daily. CsA feeding started 2 days before MERS-CoV challenge. Mice were killed 4 or 7 days post MERS-CoV infection.
RNA sequencing analysis
RNA integrity was assessed on an Experion StdSens RNA Chip (Bio-Rad Laboratories, Hercules, CA, USA). RNA-seq libraries were prepared using the TruSeq Stranded mRNA Library Prep kit (Illumina, San Diego, CA, USA). Libraries were quantified on a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and sequenced on an Illumina HiSeq 1500 platform, rapid-run mode, single-read 50 bp (HiSeq SR Rapid Cluster Kit v2, HiSeq Rapid SBS Kit v2, 50 cycles) according to the manufacturer's instructions. Quality control of RNA-seq reads was performed using the FastQC command line tool version 0.11.7. Reads were aligned using STAR version 2.7.0d to an index based on hg38 human genome version. Gene-specific read counts based on hg38 UCSC gene annotations were extracted using FeatureCounts from the Subread package version 1.6.3. Resulting read counts were imported into R. Detection of differentially expressed genes was done using DESeq2 version 1.22.1. Subsequent data analysis and visualisation was done with custom R scripts. Gene Ontology (GO) overrepresentation analysis was performed using the enrichGO function of the clusterProfiler package version 3.10.1. Sequencing data are available at Array Express, accession number E-MTAB-8222.
Statistics
All data are presented as mean±sem. Statistical significance was analysed by unpaired two-tailed t-test or by one-way ANOVA and post hoc multi-comparison tests as indicated in the respective figures. A p-value <0.05 was considered significant.
Further experimental details can be found in the supplementary material.
Results
CsA inhibits MERS-CoV replication and release in lung epithelia
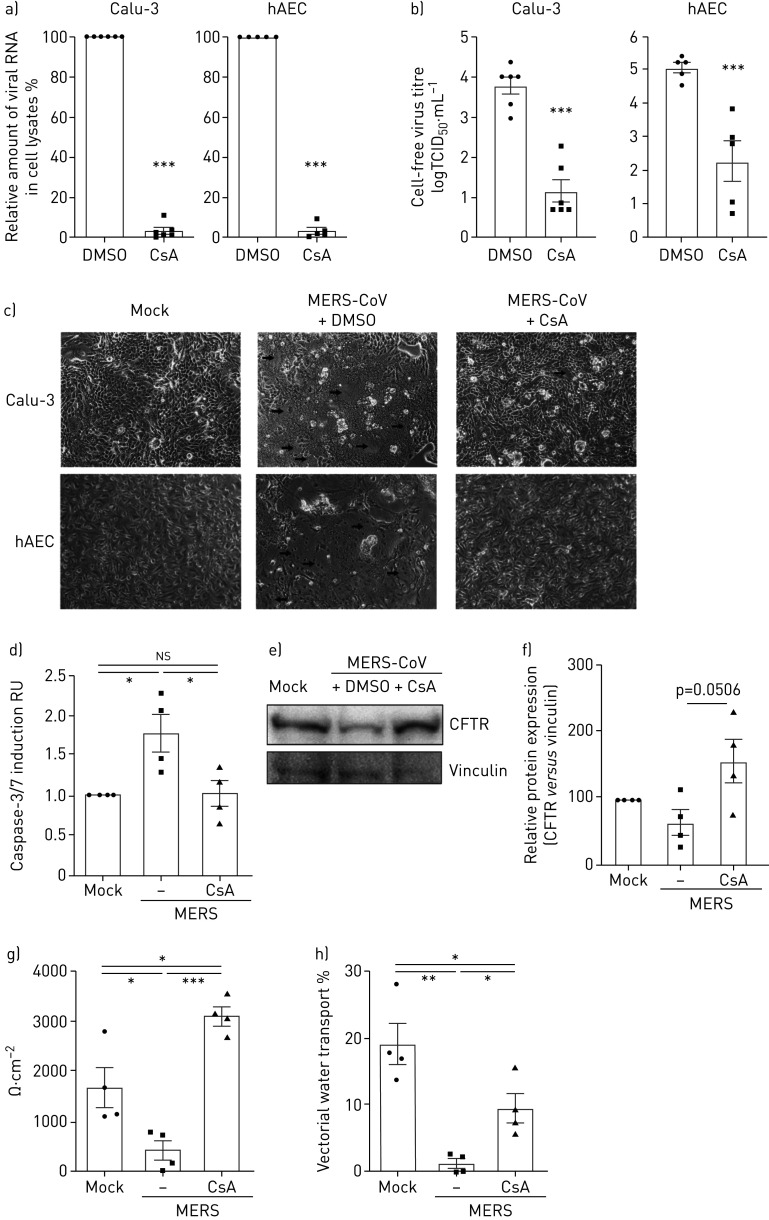
To address the previously proposed antiviral activity of CsA in clinically relevant cells, we infected the human bronchial epithelial cell line Calu-3 and primary hAECs with MERS-CoV and analysed intracellular viral RNA and infectious particle release in the presence of DMSO or CsA (figure 1). In both Calu-3 cells and hAECs, CsA treatment led to a >95% decrease of viral RNA (figure 1a) and a reduction of viral titres in the supernatant by 2.6–2.8 log10 (figure 1b). In accordance with reports from autopsy material from MERS-CoV patients [4], MERS-CoV-infected Calu-3 cells and primary hAECs both showed apoptotic cell loss and formation of multinucleated cell foci (figure 1c). Addition of CsA reduced cell foci formation and significantly reduced apoptosis induction (figure 1c, d). In line, both cystic fibrosis transmembrane conductance regulator (CFTR) (figure 1e, f) and epithelial sodium channel-β (ENaCβ) (supplementary figure E1) protein expression were improved after addition of CsA to MERS-CoV-infected Calu-3 cells. Moreover, epithelial structural integrity and vectorial water transport ability were reduced in MERS-CoV-infected control cells and significantly improved to normal levels in MERS-CoV-infected, CsA-treated cells (figure 1g, h).
FIGURE 1.
Cyclosporin A (CsA) inhibits Middle East respiratory syndrome coronavirus (MERS-CoV) replication and release in human airway epithelial cells and primary human alveolar epithelial cells (hAECs). Calu-3 cells and hAECs were infected with MERS-CoV using a multiplicity of infection of 0.1, stimulated with DMSO or 10 µM CsA, and analysed at 24 h post infection. a) Total RNA was isolated from cell lysates and viral RNA content was analysed by quantitative PCR. After normalisation to actin, changes in RNA content in CsA-treated cells were normalised to RNA present in DMSO-treated control groups (set at 100%). b) Virus titres were determined using the 50% tissue culture infective dose (TCID50) method from cell-free culture supernatant and are shown as logTCID50·mL−1. c) MERS-CoV-induced cytopathic effect and foci formation (marked by arrows) was documented in live cells by phase contrast microscopy at a magnification of ×100. d) Apoptosis induction was quantified by Caspase-3/7 Glo® Assay in Calu-3. RU: relative units. e, f) Cell lysates were analysed by Western blot for expression of cystic fibrosis transmembrane conductance regulator (CFTR) (168 kDa) and vinculin (120 kDa). e) Representative Western blots of n=3–4 experiments. f) Relative quantitation with mock samples set to 100%. g, h) Calu-3 cells were grown on transwell filters and treated as above. Epithelial integrity was quantified by transepithelial resistance measurements (g) and vectorial water transport evaluated by fluorescein isothiocyanate-dextran quantification 48 h post infection (h). Data in (a, b, d–h) are presented as mean±sem of n=3–6 experiments. Statistical significance was analysed by unpaired two-tailed t-test (a, b) and one-way ANOVA and Bonferroni's multiple comparisons test (d–h), where all groups were compared to each other. *: p<0.05; **: p<0.01; ***: p<0.005; ns: nonsignificant. Micrographs in (c) are representative of n>5 experiments.
CsA treatment affects MERS-CoV infection via CypA- and MAPK-signalling pathways
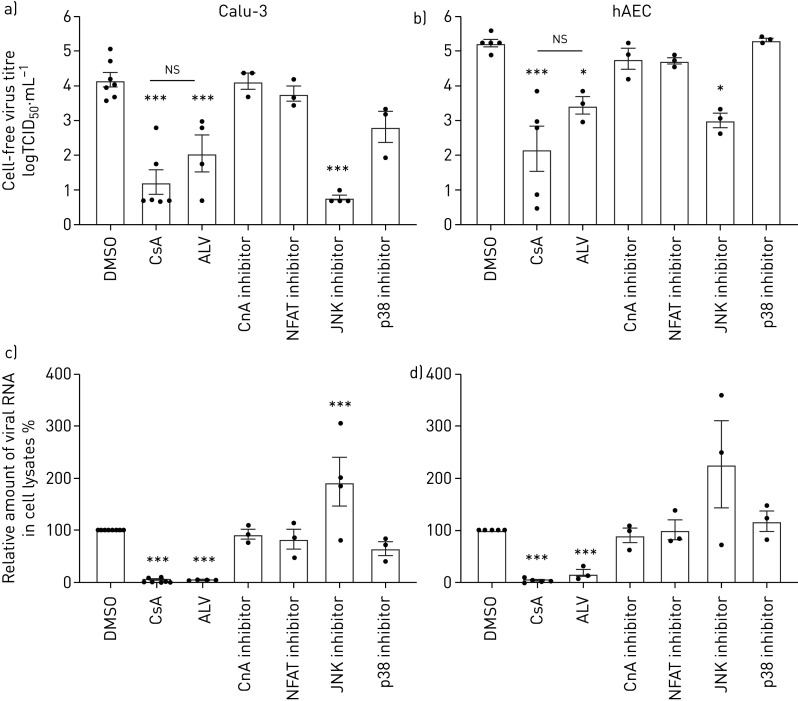
CsA is known to act via multiple signalling pathways, including cyclophilin PPIase activity, the CnA-NFAT axis and MAPK signalling [17–20]. Using specific inhibitors, we aimed to interfere with CsA-affected pathways to identify relevant molecular signalling events involved in the CsA-mediated reduction of MERS-CoV infection. Inhibition of CnA by its specific inhibitor calcineurin inhibitory peptide (CIP), as well as inhibition of the downstream transcription factor NFAT, resulted in minor, statistically nonsignificant changes in MERS-CoV viral titres in both Calu-3 cells and hAECs (figure 2a, b). The non-immunosuppressive derivate of CsA, alisporivir (ALV), which binds the PPIase but does not induce ternary complex formation of CypA with CnA, reduced viral titres by a similar extent as CsA, suggesting that it is the CypA–PPIase activity that elicits the restrictive effect on MERS-CoV replication, rather than any ternary complex-mediated signalling events. Moreover, ALV reduced cell foci formation and loss of epithelial integrity by a similar extent as CsA (supplementary figure E2). Applying specific MAPK inhibitors against JNK and p38 revealed that inhibition of JNK, but not of p38, reduced MERS-CoV titres in both Calu-3 cells and hAECs (figure 2a, b). However, inhibition of neither CnA-dependent signalling nor JNK or p38 could reproduce the CsA-induced attenuation of MERS-CoV RNA accumulation. In addition, JNK inhibition had no positive effect on cell foci formation or epithelial integrity after MERS-CoV infection (supplementary figure E3). These data suggest a role for JNK activity late in MERS-CoV replication, where adverse effects on epithelial integrity are still displayed while viral release is blocked. Of note, application of ALV resulted in a strong reduction in MERS-CoV RNA levels similar to CsA (figure 2c, d). Together, these results indicate that a CsA-induced, CypA-dependent effect has a major impact on early replication steps of MERS-CoV, strongly reducing viral RNA accumulation, even prior to virus release, independently of CnA, NFAT or JNK.
FIGURE 2.
Effect of cyclosporin A (CsA), alisporivir (ALV) and inhibition of CsA-dependent calcineurin (CnA)/nuclear factor of activated T-cells (NFATs) and mitogen-activated protein kinase (MAPK) pathways on Middle East respiratory virus-coronavirus (MERS-CoV) infection. Calu-3 cells (a, c) and human alveolar epithelial cells (hAECs) (b, d) were infected with MERS-CoV using a multiplicity of infection of 0.1. 1 h after viral adsorption, cells were stimulated with either CsA (10 µM), its non-immunosuppressive derivate ALV (10 µM), CnA inhibitor (calcineurin inhibitory peptide, 20 µM), NFAT inhibitor (50 µM), c-Jun N-terminal kinase (JNK) inhibitor (SP600125, 10 µM), p38 MAPK inhibitor (SB 203 580, 10 µM) or DMSO as solvent control. a, b) Virus titres were determined using the 50% tissue culture infective dose (TCID50) method from cell-free culture supernatant and are shown as logTCID50·mL−1. c, d) Total RNA was isolated from cell lysates at 24 h post infection and viral RNA content was analysed by quantitative PCR. After normalisation to actin, changes in RNA content in CsA-treated cells were normalised to RNA present in DMSO-treated control groups (set at 100%). Data are presented as mean±sem of n=6–8 experiments for DMSO and CsA groups and n=3–5 experiments for ALV, CnA, NFAT, JNK and p38 inhibitors. Statistical significance was analysed by one-way ANOVA and Bonferroni's multiple comparisons test, where all groups were compared to each other. Statistical significance in comparison to DMSO control is indicated. *: p<0.05; ***: p<0.005; ns: nonsignificant.
CsA treatment evokes an IFN-driven antiviral state in lung epithelial cells
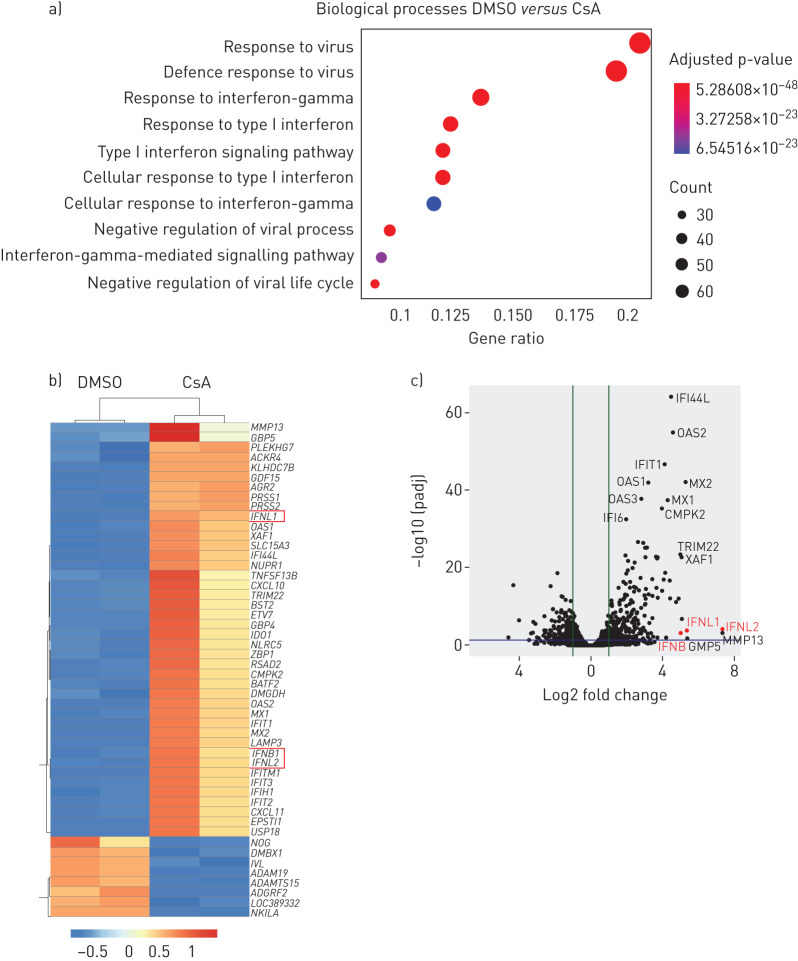
Our data suggest that, as opposed to its well-known CnA/NFAT-mediated immune-suppressive effects on immune cells, CsA might evoke an antiviral state in human lung epithelial cells. To identify the underlying mechanism, we performed RNA-seq analysis on CsA- versus DMSO-treated Calu-3 cells. Of note, analysis of enriched gene sets based on GO terms revealed that the biological processes most significantly upregulated after CsA treatment included responses to viruses and, importantly, antiviral IFN responses (figure 3a). In line with these results, both the type I IFN gene IFNB and type III IFN genes IFNL1 and IFNL2 were among the top 50 or, in the case of IFNL1, top 10 most upregulated genes in CsA-treated cells (figure 3b, c). Many of the upregulated genes were known ISGs, including MX1, MX2, OAS1, OAS2, IFIT1, IFIT2, IFIT3, LAMP3, BST2/tetherin, RSAD2/viperin and CXCL10 (figure 3b).
FIGURE 3.
Cyclosporin A (CsA) treatment induces an antiviral response state in Calu-3 lung epithelial cells. Calu-3 cells were stimulated with 10 µM CsA or treated with DMSO as vehicle control. At 24 h post stimulation, total RNA was isolated and subjected to transcriptome analysis (Methods). a) An over-representation analysis of Gene Ontology (GO) biological processes was calculated using upregulated genes with a log2 fold change >1. The 10 most significant biological processes are plotted in order of gene ratio. The size of the dots represents the number of genes in the upregulated gene list associated with the GO term; the colours of the dots represent adjusted p-values. b, c) Individual genes are shown as a heat map (b) listing the top 50 regulated genes (up and down) and as a volcano plot (c). IFNL1, 2 and 3 are highlighted in red. Data represent two independent experiments.
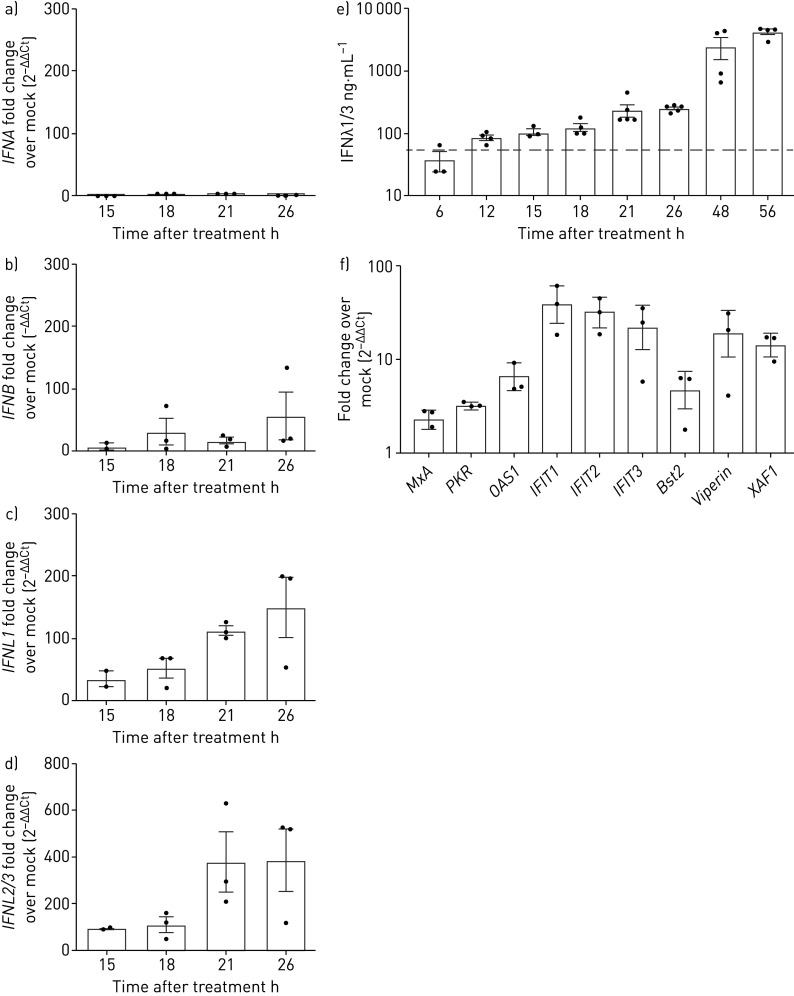
To validate our results, we analysed mRNA expression of both type I and type III IFN by quantitative real-time PCR in CsA-stimulated or DMSO-treated Calu-3 cells. We found a moderate upregulation of IFNB (up to 57-fold change over mock control) and no significant induction of IFNA (figure 4a, b). However, we revealed a strong induction of IFNL1 and IFNL2/3 mRNAs (between 150- and 387-fold change over mock control, respectively; figure 4c, d). Quantification of IFNλ1 and IFNλ3 protein release from cell culture supernatants by ELISA demonstrated a robust induction upon CsA addition as early as 12 h after CsA treatment, reaching peak values of 4222±890 ng protein·mL−1 at 48 to 56 h after CsA application (figure 4e). Similarly, treatment with ALV induced robust IFNλ release, reaching a similar maximum release of IFNλ after 72 h (supplementary figure E4). We next validated the CsA-induced upregulation of ISGs and confirmed an increased expression of selected ISGs including MxA, PKR, OAS1, IFIT1, IFIT2, IFIT3, Bst2/tetherin, RSAD2/viperin and XAF1 upon 18 h treatment with CsA compared to vehicle-treated control cells (figure 4f). These data indicate that CsA treatment mounts a distinct IFN-driven antiviral response in lung epithelial cells.
FIGURE 4.
Cyclosporin A (CsA) induces interferon-λ (IFNλ) mRNA and release. Calu-3 cells were stimulated with 10 µM CsA, followed by a–d) total RNA isolation at 15, 18, 21 and 26 h after treatment. After cDNA synthesis, quantitative PCR analysis was performed for IFNA, IFNB, IFNL1 and IFNL2/3. Fold change over DMSO-stimulated control (mock) was performed (2−ΔΔCt) to visualise IFN-induction upon CsA stimulation. e) To quantify released IFNλ, supernatants of CsA-stimulated Calu-3 cells were collected from 6 to 56 h after CsA treatment and IFNλ1/3 was determined by ELISA. f) Expression of selected IFN-stimulated genes upon CsA treatment was analysed at 18 h post CsA treatment by quantitative PCR analysis and fold change over DMSO-stimulated control (2−ΔΔCt). Data are presented as mean±sem of n=3–5 experiments in (a–e) and of n=3 experiments in (f). Grey dotted line in (e) represents the minimal detection limit of the ELISA as given by the manufacturer.
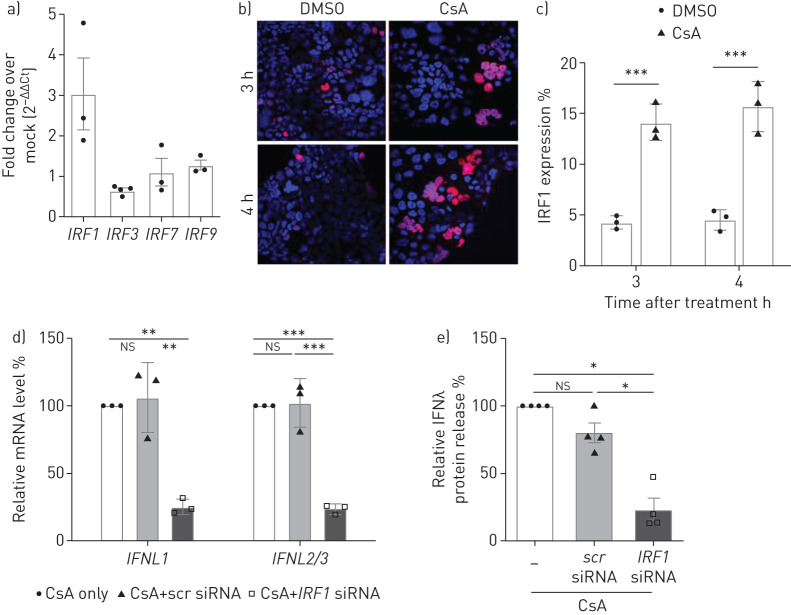
IFNλ induction is mediated by IRF1 upon CsA treatment in lung epithelial cells
To better understand the transcriptional programmes leading to IFNλ induction in CsA-treated cells, we analysed the regulation of IRFs. Our data reveal significant upregulation of IRF1 mRNA levels upon CsA treatment, but not of IRF3, IRF7 or IRF9 (figure 5a). IRF1 is known to be a specific activator of IFNL gene expression [26]. Accordingly, we identified a significantly increased number of IRF1-expressing cells in CsA-stimulated Calu-3 cells by immunofluorescence (figure 5b, c). IRF1 small interfering RNA (siRNA) knockdown significantly reduced IFNL mRNA levels in CsA-treated Calu-3 cells (figure 5d). IRF1 knockdown inhibited IFNλ release by >75% as compared to control (figure 5e).
FIGURE 5.
Interferon regulatory factor 1 (IRF1) mediates the cyclosporin A (CsA)-induced expression of interferon-λ (IFNλ). a) Calu-3 cell lysates were collected 4 h post treatment with 10 µM CsA or DMSO control. Total RNA was isolated, and quantitative PCRs for IRF1, IRF3, IRF7 and IRF9 were performed. Data are presented as fold change over DMSO (2−ΔΔCt). b) Expression of IRF1 in CsA- or DMSO-stimulated Calu-3 cells was analysed by indirect immunofluorescence on fixed and permeabilised cells. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). c) For quantification of IRF1 expression, the ratio of IRF1positive to DAPIpositive cells was calculated for at least 10 randomly chosen microscopic fields of three independent experiments. d, e) IRF1 was silenced by siRNA-transfection experiments using oligofectamine. Scrambled (scr) siRNA transfection was used as control. At 4 h post transfection, Calu-3 cells were treated with 10 µM CsA. d) Supernatants were collected 18 h after CsA treatment. IFNL mRNA was analysed via quantitative PCR and the relative fold induction in comparison to non-siRNA-treated cells (set to 100%) is shown. e) Supernatants of siRNA-transfected and CsA-stimulated cells were analysed by ELISA and the relative amount of IFNλ in comparison to non-siRNA-treated cells (set to 100%) is shown. Data are presented as mean±sem of n=3–4 experiments for (a), n=3 experiments for (b–d) and n=4 experiments for (e). Statistical significance was calculated using unpaired two-tailed t-test for (c) and one-way ANOVA and Bonferroni's multiple comparisons test comparing all groups to each other (d, e). *: p<0.05; **: p<0.01; ***: p<0.005; ns: nonsignificant.
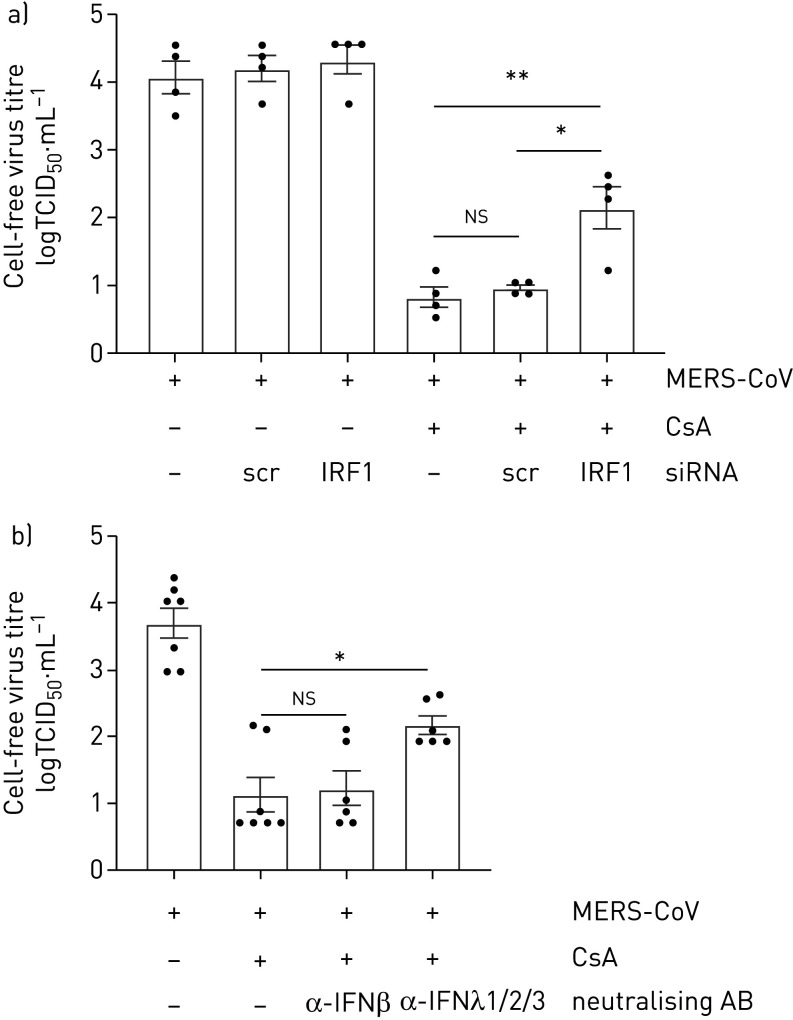
Inhibition of the IRF1–IFNλ signalling axis counteracts the MERS-CoV restrictive effect of CsA
To understand the extent to which the inhibition of MERS-CoV propagation in CsA-treated cells was mediated by IRF1-mediated production of IFNλ, we performed knockdown of IRF1 or neutralised cell-free IFNλ, respectively. Silencing of IRF1 but not treatment by control siRNA led to a significant increase in MERS-CoV-released viral particles in CsA-treated cells (figure 6a, b). Moreover, neutralising antibodies directed against IFNλ1, IFNλ2 and IFNλ3 or against the less induced IFNβ were applied (figure 6b). Neutralisation of IFNβ had no significant impact on MERS-CoV replication after CsA treatment, whereas application of anti-IFNλ1/2/3 treatment significantly increased MERS-CoV viral titres by 1.05 log10 level (figure 6b). These data indicate that the antiviral effects of CsA were at least partially mediated by an IRF1–IFNλ signalling axis, and independent of type I IFN.
FIGURE 6.
Inhibition of the interferon regulatory factor 1 (IRF1)–interferon-λ (IFNλ) signalling axis partially prevents the cyclosporin A (CsA)-induced effects on Middle East respiratory syndrome coronavirus (MERS-CoV) replication. a) IRF1 was silenced by siRNA-transfection experiments using oligofectamine. Scrambled (scr) siRNA transfection was used as control. At 20 h post transfection, Calu-3 cells were infected with MERS-CoV at a multiplicity of infection of 0.1. After 1 h of viral adsorption, cells were treated with 10 µM CsA or DMSO solvent control. 24 h post infection, viral release was determined using the 50% tissue culture infective dose (TCID50) method. b) Calu-3 cells were infected with MERS-CoV at a multiplicity of infection of 0.1 and stimulated with CsA or DMSO control as described before. In parallel, neutralising antibodies (AB) targeting IFNβ or IFNλ1/2/3 were added simultaneously with CsA. 24 h post infection viral release was determined by TCID50. Data are presented as mean±sem of n=4 experiments in (a), and n=6–7 experiments in (b). Statistical significance was calculated using one-way ANOVA and Bonferroni's multiple comparisons test, where all groups were compared to each other. Statistical significance in comparison to the oligofectamin-only treated group is indicated. *: p<0.05; **: p<0.01; ns: nonsignificant.
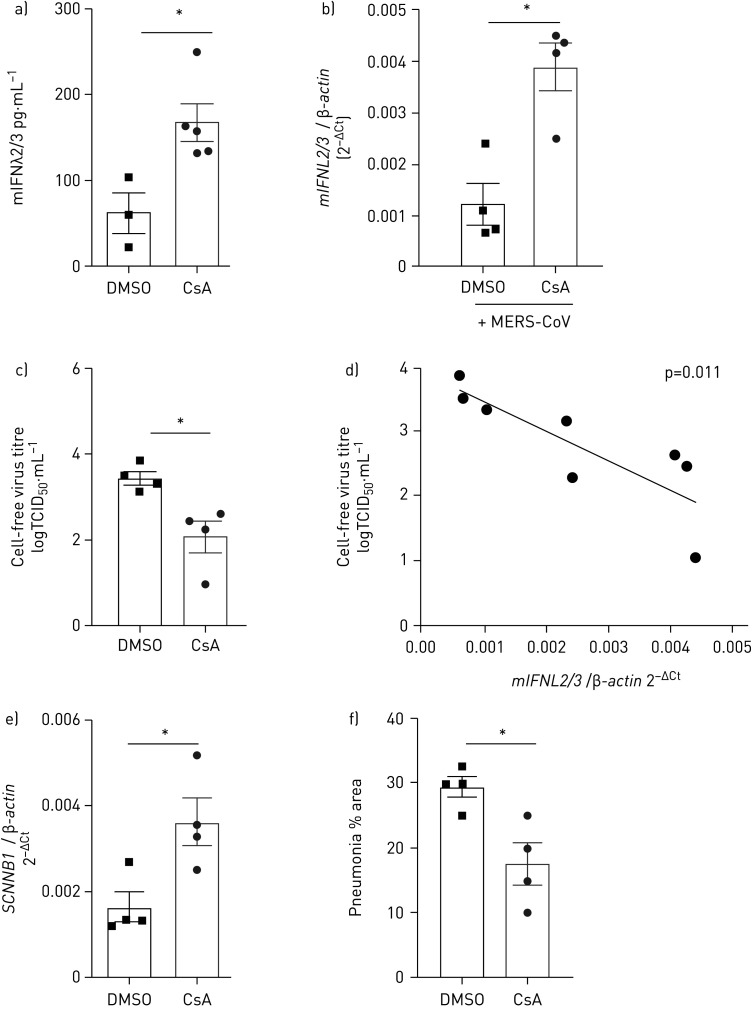
CsA treatment upregulates IFNλ and leads to reduced MERS-CoV replication and lung pathology in vivo
To validate the antiviral efficacy of CsA against MERS-CoV in vivo, we used our recently established MERS-CoV infection mouse model. This is based on the intratracheal delivery of the human DPP4 receptor to lung epithelial cells via adenoviral transduction, leading to severe MERS-CoV infection that presents as necrotising interstitial pneumonia [22]. We treated mice daily via oral intake of either DMSO or CsA, starting 2 days before mock or MERS-CoV infection. Oral CsA application resulted in CsA serum levels of 202–356 ng·mL−1 (mean 270±17 ng·mL−1), a concentration that compares to levels reached in patients under CsA treatment (supplementary figure E5 [27, 28]). Accordingly, CsA treatment significantly induced release of IFNλ in the bronchoalveolar lavage fluid (figure 7a). IFNL induction was significantly elevated in the CsA treatment group compared to the DMSO treatment group at day 7 post MERS-CoV infection (figure 7b). Oral application of CsA significantly reduced viral titres (3.45±0.15 versus 2.1±0.36 TCID50·mL−1 in the DMSO versus CsA group) at day 7 post MERS-CoV infection (figure 7c). CsA treatment did not alter adenoviral transduction efficiency (supplementary figure E6). Of note, expression levels of IFNL inversely correlated with MERS-CoV load in lung homogenates at day 7 post infection (figure 7d). A significant reduction in viral titres and a significant correlation between IFNL induction and MERS-CoV inhibition could also be demonstrated at day 4 post infection (supplementary figure E7). Expression of the SCNN1B gene (ENaCβ) as a marker of epithelial integrity was improved in lung homogenates of MERS-CoV-infected mice treated with CsA (figure 7e). While extensive oedema formation was present in a substantial proportion of MERS-CoV-infected mice, it was absent in the CsA-treated group (supplementary figure E8). Importantly, the percentage of lung area showing histopathological alterations due to MERS-CoV infection was significantly decreased by CsA treatment at day 7 post infection (figure 7f). Collectively, we have demonstrated that oral application of CsA induces IFNλ in the lungs of mice and exerts potent antiviral activity in vivo.
FIGURE 7.
Cyclosporin A (CsA) treatment upregulates interferon-λ (IFNλ) and reduces Middle East respiratory syndrome coronavirus (MERS-CoV) replication in vivo. a) Mice were fed with CsA (50 mg·kg−1·day−1) or DMSO as solvent control. After 6 days of daily oral application, levels of mouse IFNλ (mIFNλ) were analysed by ELISA from the bronchoalveolar lavage fluid. b–f) Mice were infected via the intratracheal route with an adenovirus construct encoding for human DPP4 and mCherry (Ad-hDPP4). Oral application of CsA (50 mg·kg−1·day−1) or DMSO as solvent control was started at day 3 post transduction, while mice were infected with 1.5×105 TCID50·mL−1 MERS-CoV via the intranasal route at day 5 post transduction. Mice were killed and lungs were isolated 7 days post MERS-CoV infection. Lung homogenate was used to b) quantify relative IFNL2/3 mRNA and c) determine viral load by TCID50. d) The correlation between MERS-CoV titres and IFNL2/3 mRNA expression levels was calculated using Pearson correlation efficient. e) Relative amounts of SCNNB1 (ENaCβ) RNA were determined via quantitative PCR from lung homogenates of MERS-CoV-infected and DMSO- versus CsA-treated mice. f) Lungs were fixed and processed for histological analysis. Lung areas characterised by interstitial inflammation typical for MERS-CoV-induced pneumonia [22] were quantified after whole lung processing. Statistical significance was calculated using an unpaired two-way t-test (a, b, c, d, g). *: p<0.05. TCID50: 50% tissue culture infective dose.
Discussion
With the appearance of SARS-CoV in 2002, MERS-CoV in 2012 and SARS-CoV-2 in 2019, three species of the family Coronaviridae have revealed the ability to be efficiently transmitted from human to human and to provoke serious disease with high mortality rates. Both SARS-CoV and MERS-CoV are listed on the World Health Organization blueprint list of priority diseases, and the zoonotic CoV reservoir strains are generally considered and have now been proven to be a source for emerging pandemic viruses.
Because no specific treatment is approved for MERS-CoV or SARS-CoV(-2), current treatment strategies are supportive [29, 30]. Treatments including recombinant type I IFN and antivirals (e.g. lopinavir/ritonavir) have been applied off-label to treat MERS-CoV and have yielded only moderate efficacy with controversial results in retrospective studies, and data from prospective studies or randomised controlled trials are lacking [29, 31–33]. Due to its receptor specificity to the human DPP4, to date there have been few animal models accessible for studying MERS-CoV pathogenesis and MERS-CoV-directed antiviral compounds. For this study, MERS-CoV infection in the mouse was facilitated via intratracheal delivery of a human DPP4-encoding adenovirus, which might cause low-level inflammation itself and inhomogeneous receptor distribution within the lung, present for a limited time frame. However, even if this model might not fully recapitulate the native cellular distribution or density of the receptor as seen in the human lung, high transduction efficiencies (≥95%, data not shown) allow efficient viral spread in the upper and lower respiratory airways with quick progression to severe lung injury [22] and with moderate changes in morbidity [34]. This circumvents model-specific neurotropism, as seen in some of the transgenic hDPP4 mice [35], and the necessity to adapt virus isolates via multiple passages, which might potentially affect its susceptibility to interventional strategies. While prior exposure to adenovirus evokes moderate histological changes, including perivascular and bronchiolar lymphocytic infiltration (data not shown), MERS-CoV infection leads to a clearly distinguishable granulocytic, necrotising interstitial pneumonia with alveolar oedema formation as described previously [22].
CsA has been implicated as an inhibitor of a broad spectrum of virus families, including diverse CoV [14, 36–41]. However, studies on efficacy of CsA against CoV infection have previously relied on results in liver and kidney cell lines [14–16], while results from primary lung epithelial target cells were lacking. Recently, CsA was demonstrated to restrict MERS-CoV ex vivo [13]. Still, insights on mechanistic details and on whether CsA application affects MERS-CoV infection in vivo have remained elusive.
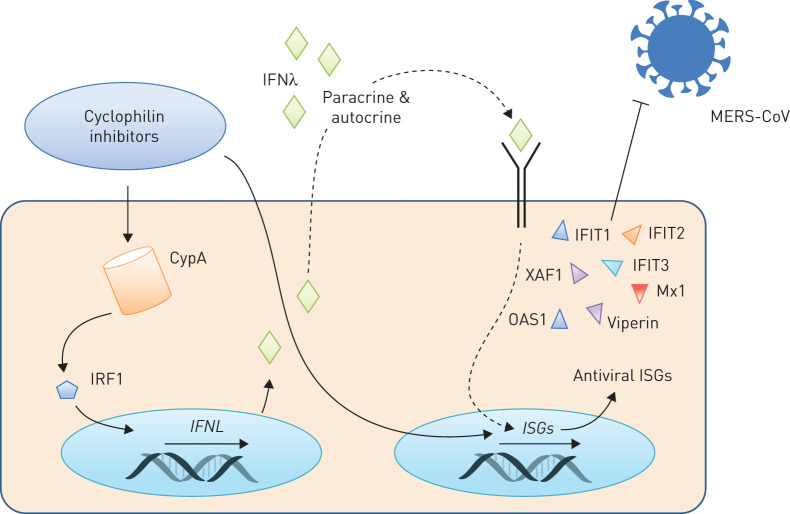
We now demonstrate that CsA application blocks MERS-CoV, both at the mRNA level and by lowering the number of infectious viral particles released, and it significantly improves epithelial barrier integrity after MERS-CoV infection. Using different inhibitors known to block CsA-targeted pathways, we have revealed that the CsA-induced blockade of MERS-CoV RNA synthesis can neither be reproduced by inhibition of known CsA-targeted MAPK nor by blockade of NFAT activation. Of note, ALV, which efficiently blocks CypA PPIase activity but affects NFAT-dependent pathways only at very high concentrations [42], diminished MERS-CoV RNA accumulation as efficiently as CsA, suggesting that CypA plays a pivotal role in these processes. In fact, we have revealed a previously unknown activation of genes involved in innate immune responses and in limitation of virus replication upon administration of CsA to lung epithelial cells. Moreover, we have demonstrated that inhibition of CypA via CsA or ALV, which both potently block CypA PPIase activity at the used concentrations [42], results in a pronounced upregulation of type III IFN at both the mRNA and protein level. This was mediated by IRF1 and was accompanied by expression of antiviral ISGs (figure 8). Among those, interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) has particularly been reported to influence the pathogenesis of MERS-CoV, highlighting the relevance of our findings [43].
FIGURE 8.
Cyclosporin A (CsA) and alisporivir (ALV) restrict Middle East respiratory syndrome coronavirus (MERS-CoV) via induction of interferon-λ (IFNλ). Cyclophilin inhibitors, including CsA or its non-immunosuppressive derivate ALV, are known modulators of cyclophilin A (CypA) and induce activation of the interferon regulatory transcription factor 1 (IRF1). Subsequently, transcription of type III interferon genes (IFNL) is upregulated and IFNλ is released both in vitro and in vivo. CsA further induces the expression of IFN-stimulated genes (ISGs), likely by auto- and paracrine signalling via IFNλ. CsA-induced ISGs include molecules with known antiviral effects, e.g. OAS1 and IFIT1, which in turn convey the restrictive effect of CsA on MERS-CoV replication.
Type III IFNs have recently emerged as key antiviral players in the innate immune response to viral infections at mucosal and epithelial surfaces [44–47]. They efficiently restrict different respiratory viruses, e.g. limiting the spread from the upper to the lower airways [44, 46–48]. As opposed to type I IFN, type III IFN do not trigger detrimental immune responses that contribute to immunopathology in influenza infection [23, 25, 44, 49]. This might prove to be pivotal in the context of CsA-dependent stimulation of IFNλ during CoV, because severe human CoV infections, e.g. MERS-CoV and, although data are still limited, SARS-CoV-2, are characterised by an immunopathology with a strong cytokine induction [5, 50, 51].
In addition to defining a novel pro-inflammatory, antiviral expression profile induced by CsA on lung epithelial cells, this study has also demonstrated for the first time that oral application of CsA reduces viral load in an in vivo MERS-CoV infection model. CsA is a licensed drug that has been in clinical use since the 1980s. While prolonged treatment (over weeks and months) with CsA can induce side effects (e.g. nephrotoxicity [48]), we here applied a short-interval oral intake of CsA during acute infection. Our results demonstrate that in vivo, oral application over 6 days results in drug serum levels that efficiently inhibit lung viral infection and pneumonia progression, highlighting CsA as a promising drug to be re-purposed for treatment of MERS-CoV.
Notably, our in vitro studies also revealed that neutralisation of type III IFNs did not completely reverse the MERS-CoV-restrictive effect of CsA. We suggest that CsA affects MERS-CoV at multiple steps during viral replication. In fact, we have shown that CsA acts on MERS-CoV propagation via inhibition of JNK, which is another downstream target of CsA [19, 20]. JNK inhibition has no impact on MERS-CoV RNA accumulation but strongly reduces the number of released infectious virions. While the exact underlying molecular mechanisms remain to be defined, this finding demonstrates that CsA likely exerts additive effects to restrict MERS-CoV replication. While application of recombinant IFNs is approved to treat virus infections and malignancies, severe side effects have been related to systemic IFN application [52]. CsA repurposing for treatment of (MERS-) CoV infection might have several advantages over IFN treatment, e.g. additional antiviral effects beyond those mediated by IFNλ alone, a favourable side-effect profile upon short-term use, a beneficial effect regarding an overshooting immune response characterising CoV disease [53, 54] and proven oral availability [55]. CsA therefore represents a promising therapeutic option to combat human CoV infections, potentially extending past MERS-CoV to the current pandemic SARS-CoV-2 strain and future CoV threats.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-01826-2019.SUPPLEMENT (771.2KB, pdf)
Shareable PDF
Acknowledgements
Work with live MERS-CoV was performed in the BSL-4 facility of the Philipps University, Marburg, Germany. We thank Julia Spengler, Larissa Hamann and Stefanie Jarmer (Dept of Internal Medicine II, University of Giessen and Marburg Lung Center (UGMLC), Giessen, Germany), and Jörg Schmidt, Cornelius Rohde, Sandro Halwe, Dirk Becker and Marc Ringel (Institute of Virology, Philipps University of Marburg, Marburg, Germany) for their excellent technical and experimental assistance. We thank Ralf Bartenschlager (Dept of Molecular Virology, University of Heidelberg, Heidelberg, Germany) for providing alisporivir.
Footnotes
This article has an editorial commentary: https://doi.org/10.1183/13993003.02484-2020
Sequencing data are available at Array Express, accession number E-MTAB-8222.
This article has supplementary material available from erj.ersjournals.com
Conflict of interest: L. Sauerhering has nothing to disclose.
Conflict of interest: A. Kupke has nothing to disclose.
Conflict of interest: L. Meier has nothing to disclose.
Conflict of interest: E. Dietzel has nothing to disclose.
Conflict of interest: J. Hoppe has nothing to disclose.
Conflict of interest: A.D. Gruber has nothing to disclose.
Conflict of interest: S. Gattenloehner has nothing to disclose.
Conflict of interest: B. Witte has nothing to disclose.
Conflict of interest: L. Fink has nothing to disclose.
Conflict of interest: N. Hofmann has nothing to disclose.
Conflict of interest: T. Zimmermann has nothing to disclose.
Conflict of interest: A. Goesmann has nothing to disclose.
Conflict of interest: A. Nist has nothing to disclose.
Conflict of interest: T. Stiewe has nothing to disclose.
Conflict of interest: S. Becker has nothing to disclose.
Conflict of interest: S. Herold has nothing to disclose.
Conflict of interest: C. Peteranderl has nothing to disclose.
Support statement: This work was supported by the German Research Foundation (KFO309 P2/P8; project ID: 284237345; SFB-TR84 B2, project ID: 114933180; SFB1021 C05, project ID: 197785619), by the German Center for Lung Research (DZL), by the German Center for Infection Research (DZIF) and the Cardio-Pulmonary Institute (CPI), EXC 2026, Project ID: 390649896. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367: 1814–1820. doi: 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV). 2018. www.who.int/en/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov) Date last accessed: January 10, 2019.
- 3.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013; 495: 251–254. doi: 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng DL, Al Hosani F, Keating MK, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol 2016; 186: 652–658. doi: 10.1016/j.ajpath.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsaad KO, Hajeer AH, Al Balwi M, et al. Histopathology of Middle East respiratory syndrome coronavirus (MERS-CoV) infection - clinicopathological and ultrastructural study. Histopathology 2018; 72: 516–524. doi: 10.1111/his.13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arabi YM, Balkhy HH, Hayden FG, et al. Middle East respiratory syndrome. N Engl J Med 2017; 376: 584–594. doi: 10.1056/NEJMsr1408795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan JF-W, Yao Y, Yeung M-L, et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis 2015; 212: 1904–1913. doi: 10.1093/infdis/jiv392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falzarano D, de Wit E, Rasmussen AL, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat Med 2013; 19: 1313–1317. doi: 10.1038/nm.3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falzarano D, de Wit E, Martellaro C, et al. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep 2013; 3: 1686. doi: 10.1038/srep01686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omrani AS, Saad MM, Baig K, et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis 2014; 14: 1090–1095. doi: 10.1016/S1473-3099(14)70920-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Tawfiq JA, Momattin H, Dib J, et al. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int J Infect Dis 2014; 20: 42–46. doi: 10.1016/j.ijid.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalhoub S, Farahat F, Al-Jiffri A, et al. IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother 2015; 70: 2129–2132. doi: 10.1093/jac/dkv085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li HS, Kuok DIT, Cheung MC, et al. Effect of interferon alpha and cyclosporine treatment separately and in combination on Middle East respiratory syndrome coronavirus (MERS-CoV) replication in a human in-vitro and ex-vivo culture model. Antiviral Res 2018; 155: 89–96. doi: 10.1016/j.antiviral.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wilde AH, Zevenhoven-Dobbe JC, van der Meer Y, et al. Cyclosporin A inhibits the replication of diverse coronaviruses. J Gen Virol 2011; 92: 2542–2548. doi: 10.1099/vir.0.034983-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbajo-Lozoya J, Ma-Lauer Y, Malešević M, et al. Human coronavirus NL63 replication is cyclophilin A-dependent and inhibited by non-immunosuppressive cyclosporine A-derivatives including alisporivir. Virus Res 2014; 184: 44–53. doi: 10.1016/j.virusres.2014.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wilde AH, Raj VS, Oudshoorn D, et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon treatment. J Gen Virol 2013; 94: 1749–1760. doi: 10.1099/vir.0.052910-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiene-Fischer C. Multidomain peptidyl prolyl cis/trans isomerases. Biochim Biophys Acta 2015; 1850: 2005–2016. doi: 10.1016/j.bbagen.2014.11.012 [DOI] [PubMed] [Google Scholar]
- 18.Banerjee S, Narayanan K, Mizutani T, et al. Murine coronavirus replication-induced p38 mitogen-activated protein kinase activation promotes interleukin-6 production and virus replication in cultured cells. J Virol 2002; 76: 5937–5948. doi: 10.1128/JVI.76.12.5937-5948.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugano N, Ito K, Murai S. Cyclosporin A inhibits collagenase gene expression via AP-1 and JNK suppression in human gingival fibroblasts. J Periodontal Res 2010; 33: 448–452. doi: 10.1111/j.1600-0765.1998.tb02343.x [DOI] [PubMed] [Google Scholar]
- 20.Matsuda S, Moriguchi T, Koyasu S, et al. T lymphocyte activation signals for interleukin-2 production involve activation of MKK6-p38 and MKK7-SAPK/JNK signaling pathways sensitive to cyclosporin A. J Biol Chem 1998; 273: 12378–12382. doi: 10.1074/jbc.273.20.12378 [DOI] [PubMed] [Google Scholar]
- 21.Volz A, Kupke A, Song F, et al. Protective efficacy of recombinant modified vaccinia virus ankara delivering Middle east respiratory syndrome coronavirus spike glycoprotein. J Virol 2015; 89: 8651–8656. doi: 10.1128/JVI.00614-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietert K, Gutbier B, Wienhold SM, et al. Spectrum of pathogen- and model-specific histopathologies in mouse models of acute pneumonia. PLoS One 2017; 12: e0188251. doi: 10.1371/journal.pone.0188251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Högner K, Wolff T, Pleschka S, et al. Macrophage-expressed IFN-β contributes to apoptotic alveolar epithelial cell injury in severe influenza virus pneumonia. PLoS Pathog 2013; 9: e1003188. doi: 10.1371/journal.ppat.1003188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malczyk AH, Kupke A, Prüfer S, et al. A highly immunogenic and protective Middle East respiratory syndrome coronavirus vaccine based on a recombinant measles virus vaccine platform. J Virol 2015; 89: 11654–11667. doi: 10.1128/JVI.01815-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peteranderl C, Morales-Nebreda L, Selvakumar B, et al. Macrophage-epithelial paracrine crosstalk inhibits lung edema clearance during influenza infection. J Clin Invest 2016; 126: 1566–1580. doi: 10.1172/JCI83931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odendall C, Kagan JC. The unique regulation and functions of type III interferons in antiviral immunity. Curr Opin Virol 2015; 12: 47–52. doi: 10.1016/j.coviro.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karamehic J, Asceric M, Tulumovic T, et al. [Serum cyclosporine levels in patients after kidney transplantation]. Med Arh 1998; 52: 131–132. [PubMed] [Google Scholar]
- 28.Narula AS, Murthy M, Patrulu K, et al. Routine cyclosporine concentration - C2 level monitoring. Is it helpful during the early post transplant period? Med J Armed Forces India 2004; 60: 326–328. doi: 10.1016/S0377-1237(04)80003-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zumla A, Chan JFW, Azhar EI, et al. Coronaviruses — drug discovery and therapeutic options. Nat Rev Drug Discov 2016; 15: 327–347. doi: 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–1720. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017; 9: eaal3653. doi: 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arabi YM, Alothman A, Balkhy HH, et al. Treatment of Middle East respiratory syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials 2018; 19: 81. doi: 10.1186/s13063-017-2427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Tawfiq JA, Memish ZA. Update on therapeutic options for Middle East respiratory syndrome coronavirus (MERS-CoV). Expert Rev Anti Infect Ther 2017; 15: 269–275. doi: 10.1080/14787210.2017.1271712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Li K, Wohlford-Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA 2014; 111: 4970–4975. doi: 10.1073/pnas.1323279111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal AS, Garron T, Tao X, et al. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 2015; 89: 3659–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma C, Li F, Musharrafieh RG, et al. Discovery of cyclosporine A and its analogs as broad-spectrum anti-influenza drugs with a high in vitro genetic barrier of drug resistance. Antiviral Res 2016; 133: 62–72. doi: 10.1016/j.antiviral.2016.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nkongolo S, Ni Y, Lempp FA, et al. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol 2014; 60: 723–731. doi: 10.1016/j.jhep.2013.11.022 [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa M, Sakamoto N, Tanabe Y, et al. Suppression of hepatitis C virus replication by cyclosporin A is mediated by blockade of cyclophilins. Gastroenterology 2005; 129: 1031–1041. doi: 10.1053/j.gastro.2005.06.031 [DOI] [PubMed] [Google Scholar]
- 39.Karpas A, Lowdell M, Jacobson SK, et al. Inhibition of human immunodeficiency virus and growth of infected T cells by the immunosuppressive drugs cyclosporin A and FK 506. Proc Natl Acad Sci USA 1992; 89: 8351–8355. doi: 10.1073/pnas.89.17.8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Shi Y, Qi J, et al. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J Virol 2013; 87: 5949–5958. doi: 10.1128/JVI.00545-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Z, He H, Wu Y, et al. Cyclosporin A inhibits rotavirus replication and restores interferon-beta signaling pathway in vitro and in vivo. PLoS One 2013; 8: e71815. doi: 10.1371/journal.pone.0071815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallay PA. Cyclophilin inhibitors: a novel class of promising host-targeting anti-HCV agents. Immunol Res 2012; 52: 200–210. doi: 10.1007/s12026-011-8263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menachery VD, Gralinski LE, Mitchell HD, et al. Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere 2017; 2: e00346-17. doi: 10.1128/mSphere.00346-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davidson S, McCabe TM, Crotta S, et al. IFNλ is a potent anti-influenza therapeutic without the inflammatory side effects of IFNα treatment. EMBO Mol Med 2016; 8: 1099–1112. doi: 10.15252/emmm.201606413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazear HM, Nice TJ, Diamond MS. Interferon-λ: immune functions at barrier surfaces and beyond. Immunity 2015; 43: 15–28. doi: 10.1016/j.immuni.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klinkhammer J, Schnepf D, Ye L, et al. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife 2018; 7: e33354. doi: 10.7554/eLife.33354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamming OJ, Terczyńska-Dyla E, Vieyres G, et al. Interferon lambda 4 signals via the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J 2013; 32: 3055–3065. doi: 10.1038/emboj.2013.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baños-Lara MDR, Harvey L, Mendoza A, et al. Impact and regulation of lambda interferon response in human metapneumovirus infection. J Virol 2015; 89: 730–742. doi: 10.1128/JVI.02897-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galani IE, Triantafyllia V, Eleminiadou E-E, et al. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 2017; 46: 875–890. doi: 10.1016/j.immuni.2017.04.025 [DOI] [PubMed] [Google Scholar]
- 50.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 2017; 39: 529–539. doi: 10.1007/s00281-017-0629-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis 2020; 71: 762–768. doi: 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lasfar A, Zloza A, Cohen-Solal KA. IFN-lambda therapy: current status and future perspectives. Drug Discov Today 2016; 21: 167–171. doi: 10.1016/j.drudis.2015.10.021 [DOI] [PubMed] [Google Scholar]
- 53.DeDiego ML, Nieto-Torres JL, Regla-Nava JA, et al. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol 2014; 88: 913–924. doi: 10.1128/JVI.02576-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol 2020; 92: 424–432. doi: 10.1002/jmv.25685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giordanetto F, Kihlberg J. Macrocyclic drugs and clinical candidates: what can medicinal chemists learn from their properties? J Med Chem 2014; 57: 278–295. doi: 10.1021/jm400887j [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-01826-2019.SUPPLEMENT (771.2KB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-01826-2019.Shareable (332.8KB, pdf)