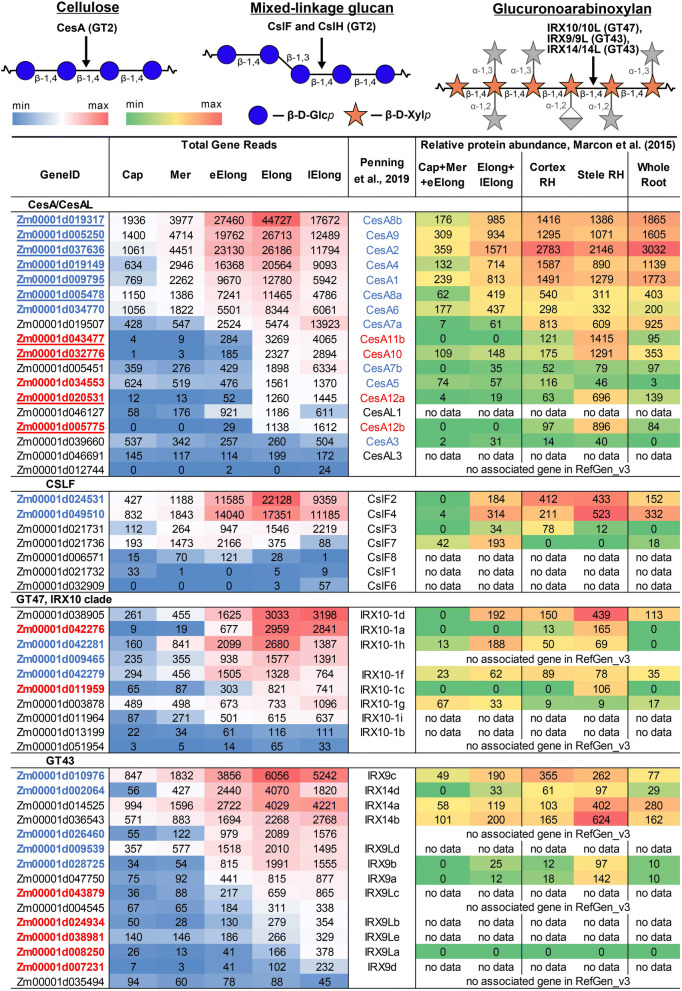
Figure 3.
Expression level (TGR, red-blue heat map) and relative protein abundance (averaged and normalized total spectral counts22, red-green heat map) of ZmCesA/CesAL, ZmCslFs and genes encoding members of the xylan backbone synthase complex in various zones of maize root. Heat map color coding is applied separately to each gene subgroup. The underlined gene names indicate the baits for co-expression analysis. The genes co-expressed with maize primary cell wall CesAs are labelled in blue, and genes co-expressed with secondary cell wall CesAs are labelled in red. Annotations are based on the study by Penning et al.18, and are obtained by matching of the RefGen_v3 and RefGen_v4 gene models. The annotations shown in blue and in red are CesAs assigned to primary and secondary cell wall formation, respectively, by Penning et al.18. Cap—root cap, Mer—meristem, eElong—early elongation zone, Elong—zone of active elongation, lElong—zone of late elongation before root hair initiation, and RH—root hair zone. No data, i.e., no corresponding peptides were obtained from any of the studied root samples22.