Figure 1: Pervasive increases in endophytic bacterial load in herbivore-damaged leaves.
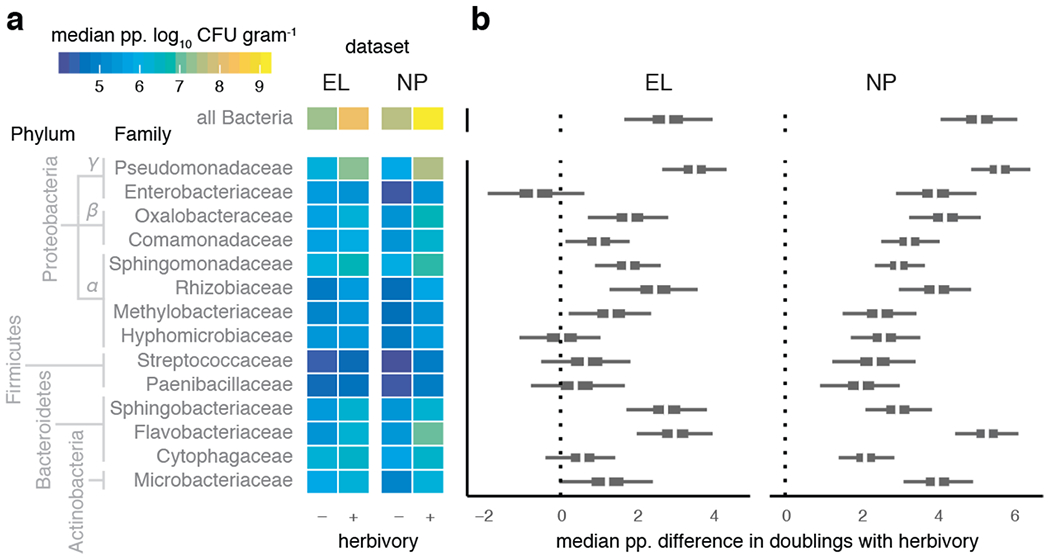
a–b. Posterior predicted (‘pp.’) infection intensity of bacterial amplicon sequence variants (bASV) from the 14 most prevalence bacterial families show variation in the extent of elevated growth in herbivore-damaged leaf tissue. a. Heatmap shows median predicted log10 bacterial abundance (colony-forming units, CFU) per g starting leaf material) from 200 posterior simulations of the best-fitting model of each bacterial family separately (see Methods). b. Boxplots showing median (white), 95% (thin), and 50% (thick) quantiles of the posterior predicted median difference in the number of bacterial cell divisions (i.e., doublings) achieved in herbivore-damaged leaves compared to undamaged leaves, for sites EL and NP separately.
