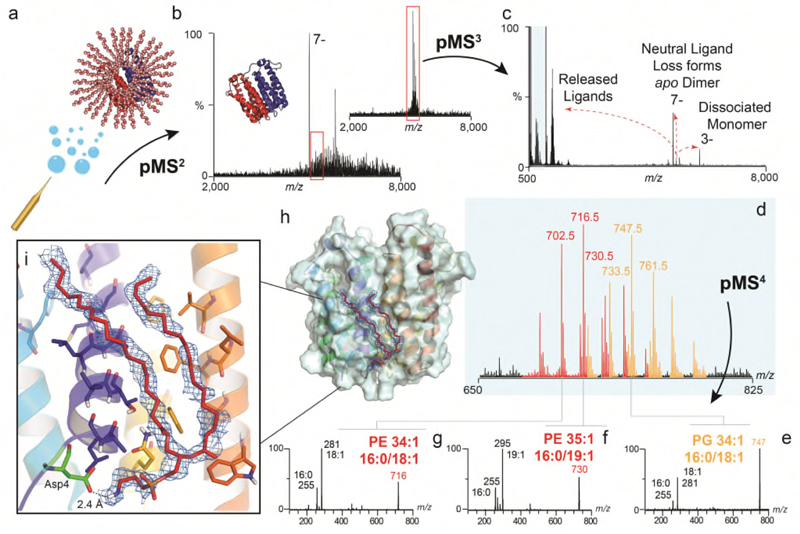
Figure 3. Identification of unknown ligands bound to TSPO and subsequent fitting of PE 16:0/18:1 into unresolved electron density in the X-ray structure.
(a) Schematic showing electrospray and release of the TSPO dimer from detergent micelles in negative ion polarity (subunits red and blue cartoon, with detergent micelle orange spheres) (b) Native mass spectrum of the TSPO dimer (pMS2) and isolation of the 7- charge state (red box) (inset). (c) Collisional activation (pMS3) yields dissociated monomer, apo dimer produced from neutral ligand loss, and multiple ligands at low m/z (blue box). (d) Zoom of low m/z region showing two peak series corresponding to multiple lipids PE (red) and PG (orange). (e-g) Isolation and subsequent fragmentation of released lipids (pMS4) defines the hydrocarbon chain length and extent of unsaturation. (h) Fitting of the most abundant PE lipid identified - PE (16:0/18:1) (red sticks) into the electron density (blue mesh) in the TSPO A139T crystal structure (each monomer in blue and yellow helices) (PDB 4UC1). Critical protein-lipid interactions are shown (zoom box) with PE interacting favourably with surrounding amino acids through hydrophobic interactions and the headgroup terminal amine forming a hydrogen-bonded interaction with the carboxylic acid side chain of Asp4 (green sticks). Other homologous PEs with different acyl tails (e.g. 16:0/19:1 and 18:/18:1) can also be accommodated within this density. See Online Methods for MS parameters and Supplementary Figure 9 for complete MS dataset. Lipid identification is representative of two independent protein preparations.