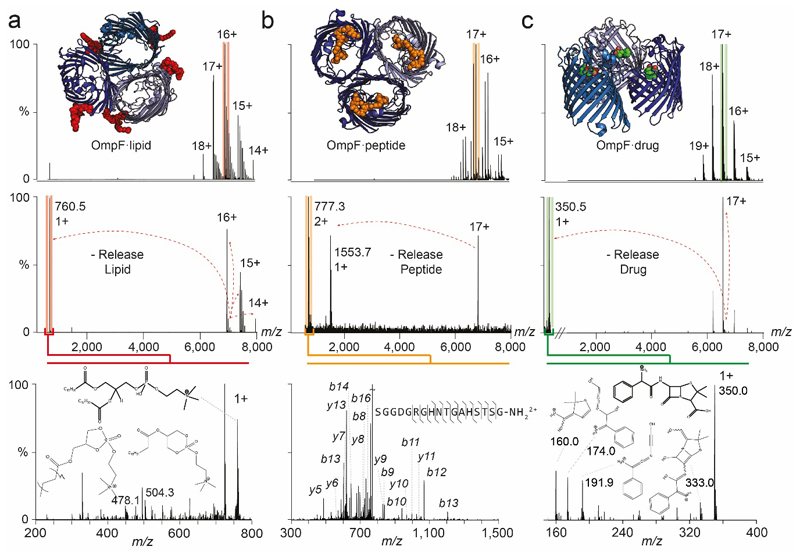
Extended Data Figure 4. Nativeomics defines lipid, peptide and drug bound to the trimeric membrane porin OmpF through progressive dissection using multiple stages of MSn.
(a) OmpF bound to lipids is released from detergent micelles (pMS2), the 16+ charge state isolated (red), with lipids released from the complex (pMS3), the peak (m/z 760.5) is then isolated and fragmented to yield fragments at m/z 504.3 and 478.1 (MS4). Spectral matching assigns this lipid as PC 16:0/18:1 (b) OmpF bound to peptide OBS1 is released from detergent micelle (pMS2) and a charge state assigned to the OmpF trimer and three OBS1 peptides was isolated (17+, orange) for pMS3. m/z 777.3 is isolated and fragmented (pMS4) to yield b and y ions that enable peptide sequence determination. (c) OmpF bound to ampicillin. The 17+ charge state (green) was isolated for activation and the ligand released at m/z 350.5 (pMS3). Fragmentation (pMS4) yields characteristic ions that identify ampicillin following database searching or spectral matching. For MS parameters see Online Methods.