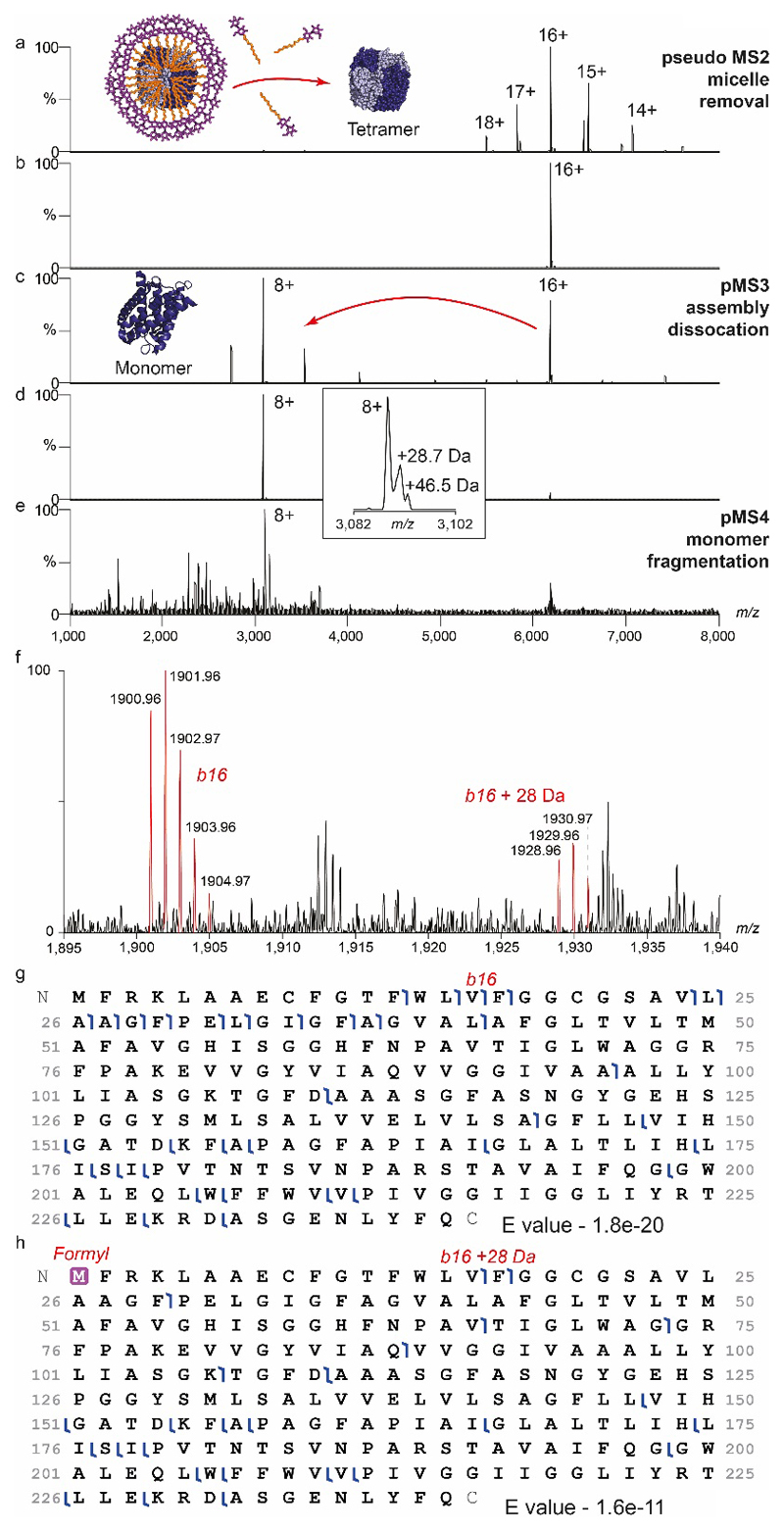
Extended Data Figure 5. Nativeomics applied to AqpZ tetramer to progressively dissect the assembly and identify multiple proteoforms.
(a) pMS2 effects the removal of the C8E4 detergent micelle in the source region (in-source activation 136V, source compensation voltage 10%) (b) the 16+ charge state (m/z 6,183) is then selected using the ion trap and (c) dissociated (MS3) in the HCD cell (NCE 22%) ejecting monomers. (d) Isolation of the monomer (8+) (m/z 3,090) is performed in the ion trap, the inset (expansion) shows additional species at ~ +29 Da and ~ +46 Da. (e) pMS4 top-down fragmentation (CID NCE 20%) of these species in the ion trap and detection at high-resolution in the Orbitrap reveals predominantly b and y fragment ions in the low m/z region. (f) Assignment of the 1+ charge, b16 ion at m/z 1,900.96, and +28 Da partner at m/z 1,928.96 is consistent with assignment of the +28 Da modification to N-terminal formylation as previously suggested15. Percentage formylation of 30-40% estimated based on b ion intensity ratios. (g) Coverage of non-modified AqpZ identified with E value of 1.8e-20. h) Coverage map of N-terminally formylated AqpZ identified with E value of 1.6e-11.
The top-down data presented here, does not represent a fully optimised top-down experiment for proteoform ID. Longer acquisition times and other complimentary fragmentation methods would likely improve sequence coverage, however, this data clearly demonstrates the capability of the modified ion trap on the Orbitrap Eclipse platform for isolation and fragmentation of high m/z ions (m/z 3,000-8,000). Furthermore it showcases the application of protein-centric Nativeomics to isolate individual membrane protein assemblies from mixtures, dissect them into subunits and identify proteoforms through top-down pMS4. Data is representative of two biological repeats.