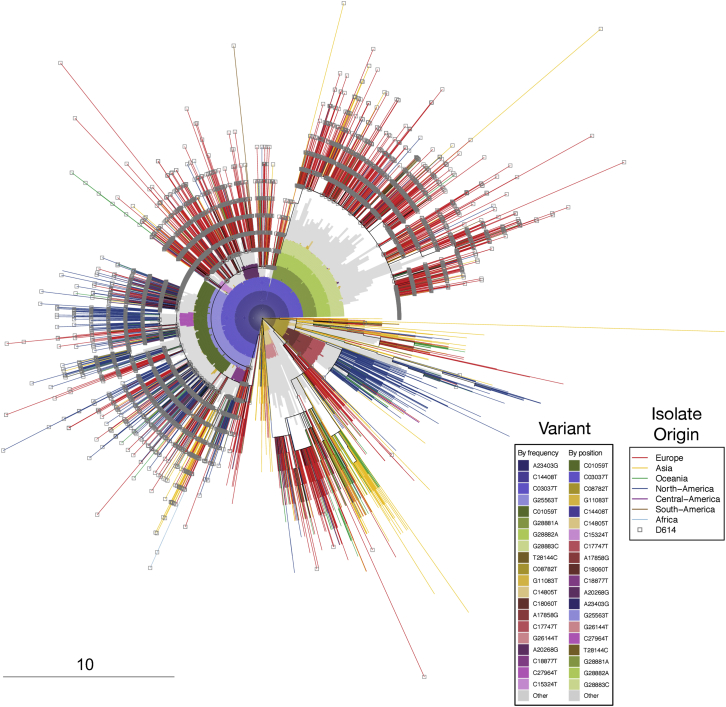
Figure S6.
Distribution of A23403G (D614G) Mutation and Other Mutations on an Approximate Phylogenetic Tree Using Parsimony, Related to Figure 7
This tree the same as the tree shown in Figure 6A, but highlights complementary information: the G614 substitution, and patterns of bases that underly the clades. It is based on the “FULL” alignment of 17,760 sequences, from the June 2 alignment, described in the pipeline in Figure S1, from the beginning of (orf1ab) to the last stop-codon (ORF10), NC_045512 bases 266-29,674). The outer element is a radial presentation of a full-coding-region parsimony tree; branches are colored by the global region of origin for each virus isolate. The inner element is a radial bar chart showing the identity of common mutations (any of the top 20 single-nucleotide mutations from the June 2 alignment), so that sectors of the tree containing a particular mutation at high frequency are subtended by an inner colored arc; mutations not in the top 20 are presented together in gray. The tree is rooted on a reference sequence derived from the original Wuhan isolates (GenBank accession number NC_045512), at the 3 o’clock position. Branch ends representing sequence isolates bearing the D614G change are decorated with a gray square; sectors of the tree containing that mutation are subtended by a dark blue arc in the inner element; other mutations are denoted by different colors. As an example, in this tree, the region from approximately 12:30 to 3 o’clock represents GISAID’s “GR” clade, defined both by mutations we are tracking in this paper that carry the G614 variant (the GISAID G clade, defined by mutations A23403G, C14408T, C3037T, and a mutation in the 5′ UTR (C241T, not shown here), and an additional 3-position polymorphism: G28881A + G28882A + G28883C. These base substitutions are contiguous and result two amino acid changes, including N-G204R, hence GISAID’s “GR clade” name. Close examination of this triplet in sequences from the Sheffield dataset suggests the mutations are not a sequencing artifact. The outer phylogenetic tree was computed using oblong (see STAR Methods), and plotted with the APE package in R. The inner element is a bar chart plotted with polar coordinates using the gglot2 package in R. The frequency of the GR clade appears to be increased in the UK and Europe as a subset of the regional G clade expansion, given that both carry G614D.