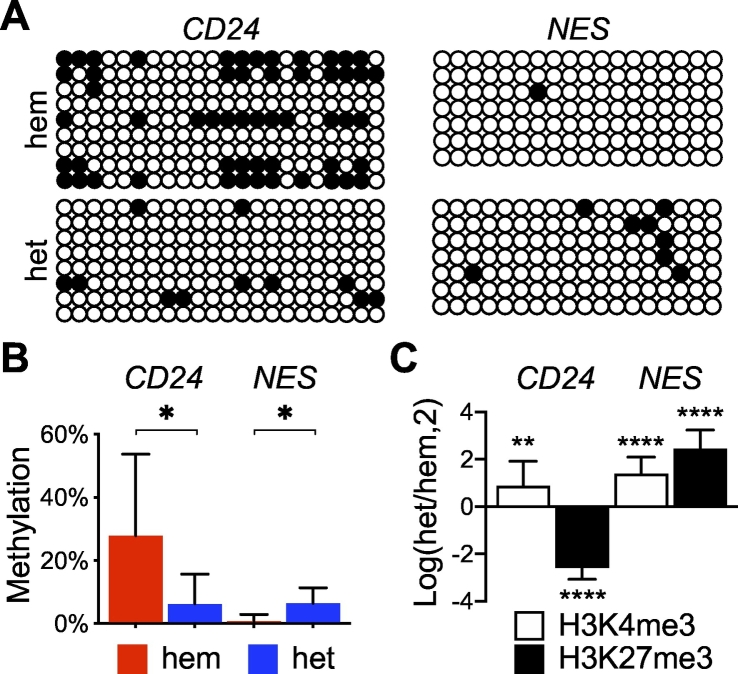
Fig. 6.
Differential epigenetic modifications between CD24 and NES in IDH1R132H-heterozygous cells. A, Bisulfite sequencing results showing lower levels of CpG methylation (filled circles) in CD24 but higher levels in NES in IDH1R132H-heterozygous cells than in IDH1R132H-hemizygous cells. B, Quantitative analysis of the bisulfite sequencing results. C, Chromatin immunoprecipitation of BT142 spheroids revealed an opposing trend of H3K27me3 (n = 9) between CD24 and NES, presented in a log2 ratio of IDH1R132H-heterozygous cells over IDH1R132H-hemizygous cells.