Fig. 1.
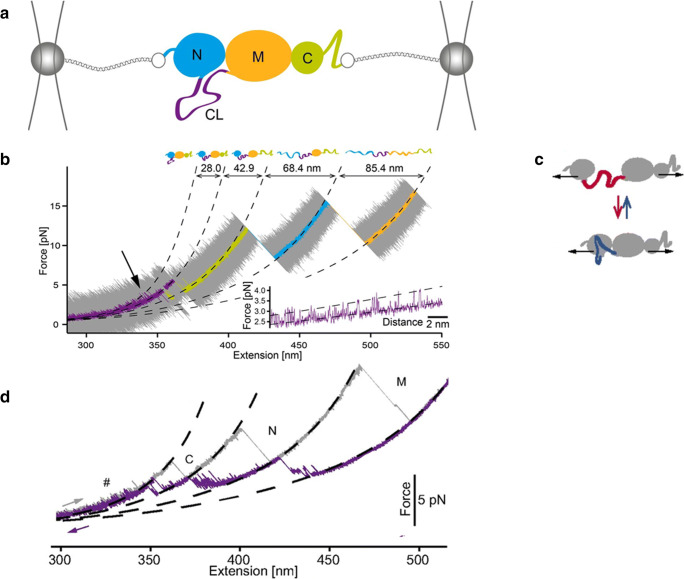
Experimental set-up used to manipulate single Hsp82 monomers with optical tweezers (Jahn et al. 2014; Jahn et al. 2016; Jahn et al. 2018). a The monomer comprises a N-terminal ATP–binding domain (N, 211 residues), a middle domain (M, 266 residues) responsible for client binding and a C-terminal domain (C, 172 residues) responsible mainly for dimerization. The N- and M-domains are connected to each other by a charged linker (CL), shown in purple. For mechanical manipulation, the protein is attached between two glass beads by means of two DNA molecular handles. By varying the position of one of the two traps, the force applied on Hsp82 monomer can be varied. b Stretching trace obtained by pulling on a Hsp82 monomer at constant speed. The original data (20 kHz) were filtered to obtain smooth a trace showing three major transitions (rips) corresponding to the sequential unfolding of the C-, N- and M-domain, respectively. The inset shows fluctuations of Hsp82 monomer at low forces due to hopping of the CL between a compact and an extended conformation, as schematically shown in panel c). d Stretching (in grey) and relaxation (in purple) cycle acquired by pulling and relaxing a Hsp82 monomer. The relaxation trace shows various fluctuations corresponding to an ensemble of intermediate states. Reprinted with permission from PNAS, (Jahn et al. 2014; Jahn et al. 2016)