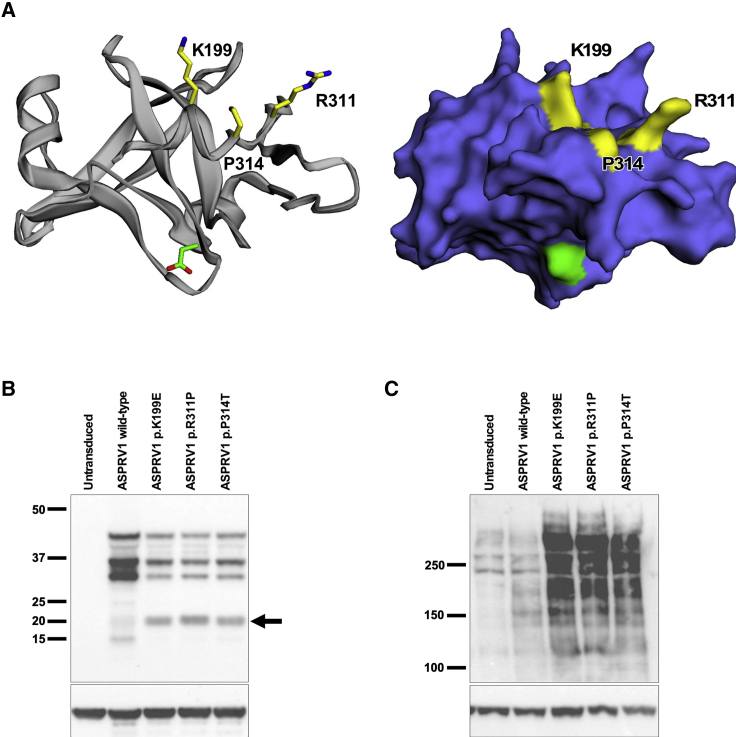
Figure 3.
Clustered ASPRV1 Mutations Generate a Novel ASPRV1 Auto-cleavage Product and Result in Accumulation of High-Molecular-Weight Forms of Filaggrin
(A) A model of the ASPRV1 active enzyme crystal structure9 (ribbon and stick rendering on left, surface rendering on right) shows that all three pathogenic mutation sites (yellow) are tightly clustered within the three-dimensional protein. The catalytic aspartate is shown in green.
(B and C) Human keratinocytes (HK), either nontransduced or transduced with wild-type or mutant ASPRV1, were switched to high calcium (1.2 mM) for 8 days to promote differentiation. Immunoblotting of protein extracts was performed with anti-actin (Sigma SAB4200248, 1:2,000) as a loading control on replicate blots to show equivalent protein loading across all samples. (B) Immunoblotting with an anti-HA antibody (Cell Signaling 3724, 1:500) shows bands at ~37 kDa and ~28 kDa in cells expressing either wild-type or mutant forms of HA-tagged ASPRV1. In addition, cells expressing each of the ASPRV1 mutant forms show a unique ~20 kDa band not seen in nontransduced cells or cells expressing wild-type ASPRV1 (arrow). (C) Immunoblotting with an anti-filaggrin antibody directed against the N-terminal domain (residues 1–261, Thermo Fisher Scientific PA5-79267, 1:500) shows profilaggrin and higher molecular weight filaggrin cleavage products within differentiated keratinocytes. In nontransduced keratinocytes and keratinocytes transduced with wild-type ASPRV1, there is a low quantity of high-molecular-weight filaggrin products, consistent with normal processing of filaggrin to filaggrin repeat monomers. Cells expressing mutant ASPRV1 (encoding p.Lys199Glu, p.Arg311Pro, or p.Pro314Thr) demonstrate an accumulation of high-molecular-weight filaggrin products, suggesting that ASPRV1 mutations impair filaggrin processing.