Figure 4.
Structural Basis of SCN5A Loss-of-Function Variants
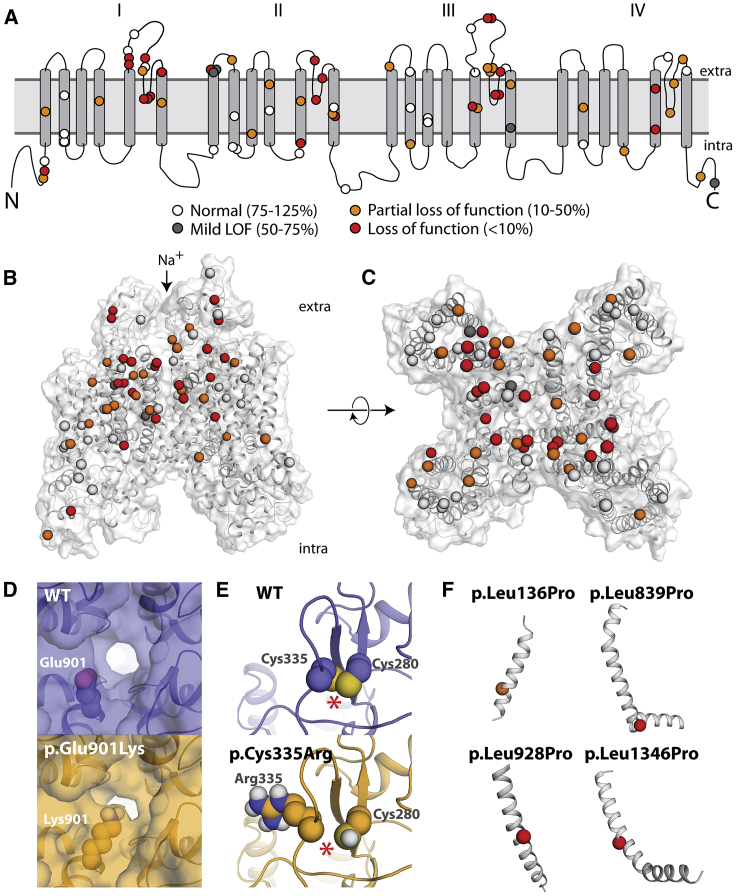
(A) Two-dimensional schematic of NaV1.5 structure. All previously unstudied variants are shown and color-coded based on peak current density (white 75%–125%, gray 50%–75%, orange 10%–50%, red < 10%).
(B and C) Three-dimensional homology model of NaV1.5. Variants are colored as in (A).
(D) Top-down view of WT (top) and p.Glu901Lys (bottom), as modeled using Rosetta. The lysine residue projects into the pore, likely disrupting sodium passage.
(E) View of WT and p.Cys335Arg, as modeled using Rosetta. The WT protein has a disulfide bond between Cys335 (left) and Cys280 (right), which was inferred from the spatial proximity of these two residues and the fact that the corresponding residues in the template structures also form a disulfide bond; this bond is disrupted by p.Cys335Arg. The disulfide bond is indicated with an asterisk (∗).
(F) Four leucine -> proline variants in this study. p.Leu136Pro is a partial loss-of-function variant and p.Leu839P, p.Leu928Pro, and p.Leu1340Pro are loss of function. The structures of these four variants were not modeled because modeling drastic structural changes involving prolines that are part of a helix usually cannot be reliably modeled in Rosetta. However, these variants likely cause loss of protein function by causing a kink in the alpha helix and protein misfolding.