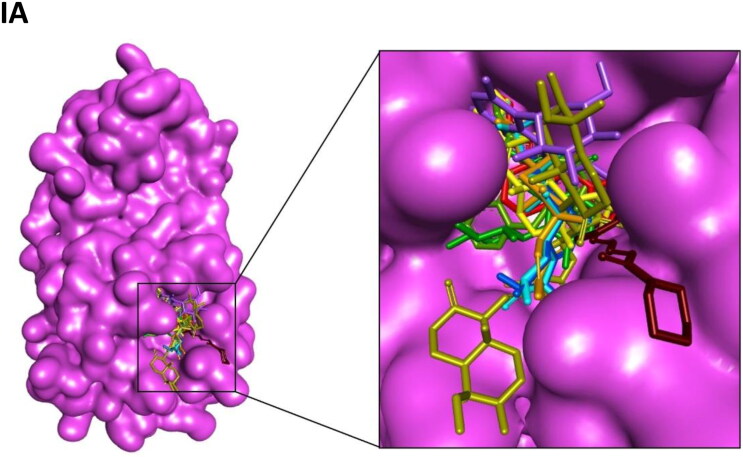
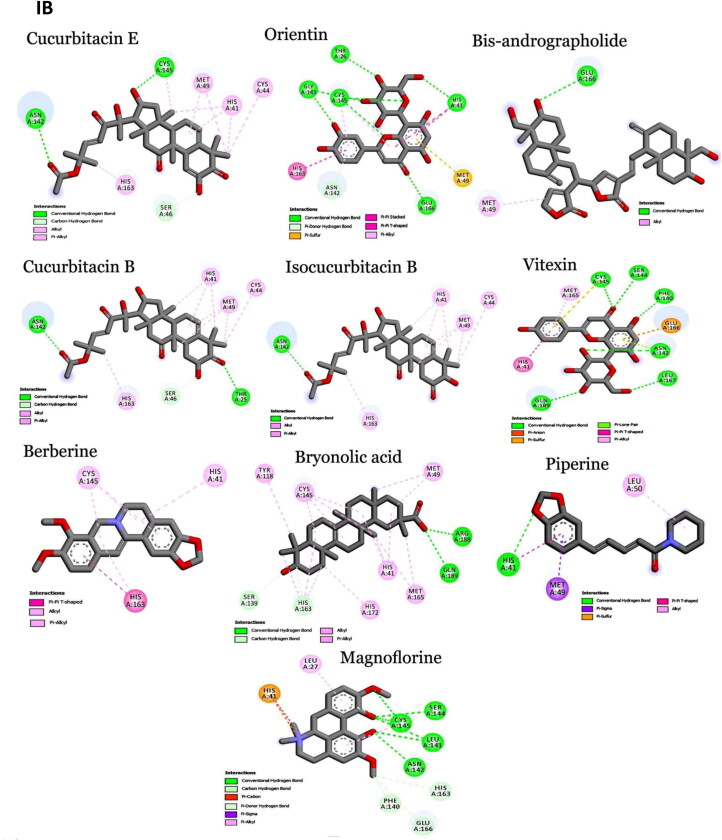
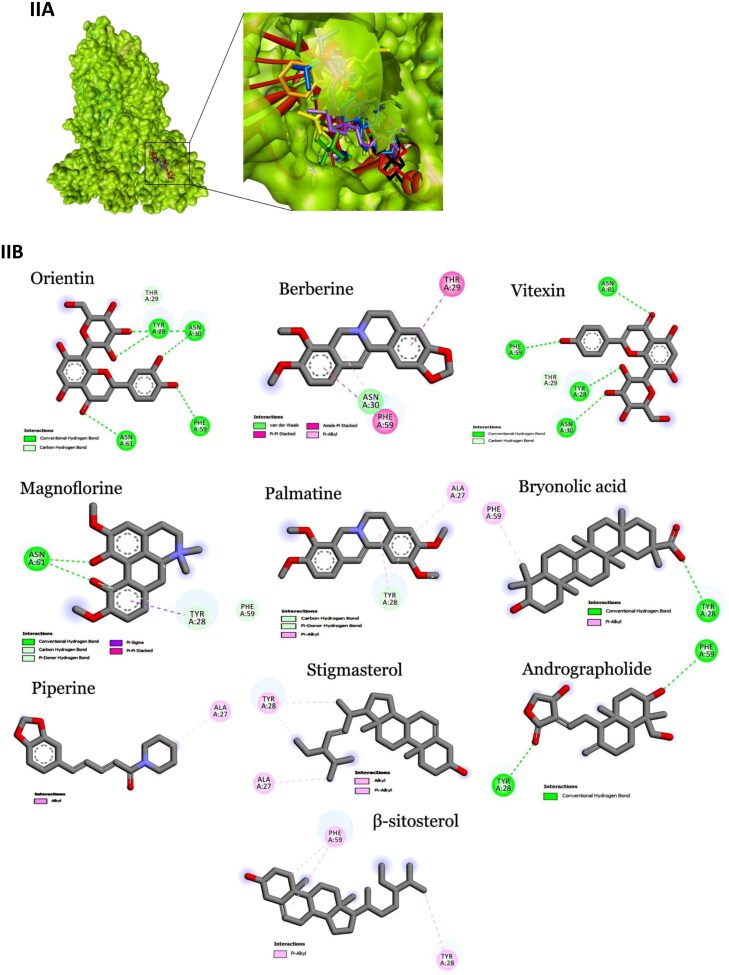
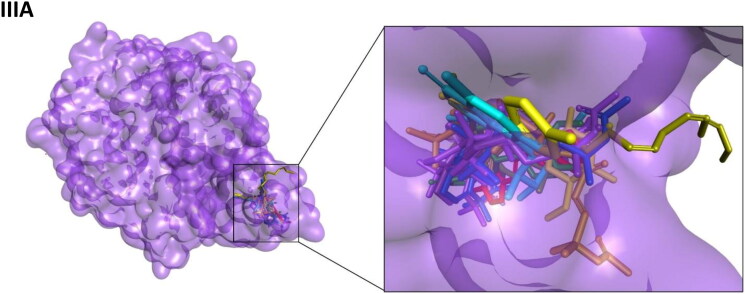
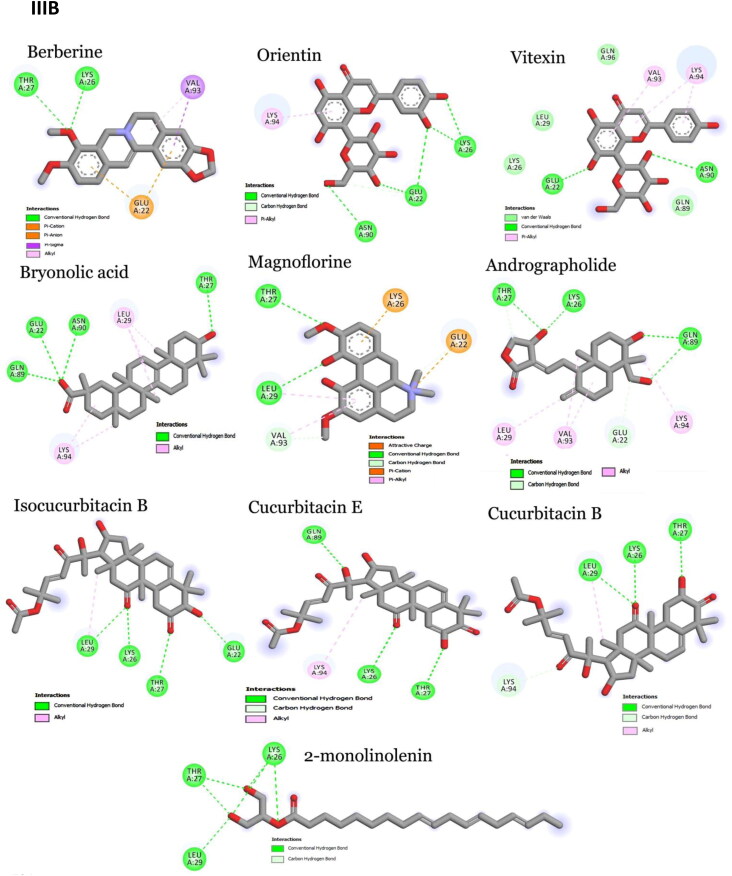
Figure 2.
Interaction map of top ten scoring ligands with SARS-CoV-2 drug targets (I) Main protease (Mpro, 5R82) (II) Spike protein (6VYB) (III) ACE-2 (1R42) (A) Crystal structure of drug targets (represented in space-filling model) displaying the druggable pocket of ligands (stick model) (B) 2D interaction map showing the top ten ligand interactions with receptor. Active site binding pockets residues are represented in three letter amino acid code and the type of interactions are mentioned in different colors.