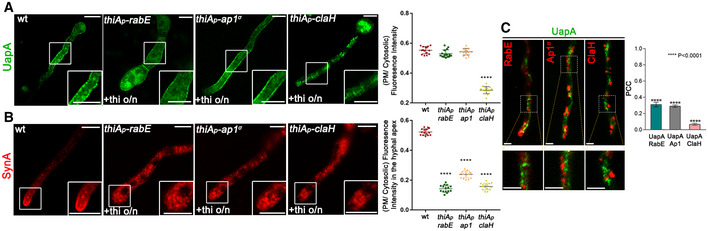
Figure 5. UapA translocation to the PM takes place without involvement of conventional post‐Golgi machinery.
-
AEpifluorescence microscopy analysis examining the subcellular localization of de novo made UapA‐GFP, after 6–8 h of initiation of transcription in strains where the expression of RabE, AP‐1σ or ClaH (thiA p ‐rabE, thiA p ‐ap1 σ or thiA p ‐claH) has been repressed ab initio (o/n) by addition of thiamine. Notice that when rabE or ap1 σ is repressed UapA‐GFP still reaches the PM. In contrast, repression of claH abolishes labelling of the PM and leads to the cytosolic puncta and a membranous network. Scale bars: 5 μm. Quantification: UapA‐GFP PM/cytosolic intensity ratios are plotted on the right. Mean ratios for wild type, thiA p ‐rabE, thiA p ‐ap1 σ and thiA p ‐claH are 0.55 ± 0.02, 0.53 ± 0.03, 0.54 ± 0.02 and 0.29 ± 0.02, respectively. For the statistical analysis, Tukey's multiple comparison test was performed (one‐way ANOVA). After 6‐8 h of transcriptional derepression, UapA‐GFP fluorescence to the PM does not change when rabE or ap1 σ are repressed. On the other hand, in the absence of ClaH, UapA does not reach the PM, as its fluorescence intensity there is statistically lower (****P < 0.0001) in comparison with that of the wild‐type strain. Biological/technical replicates: 2/15 for each strain.
-
BIn a similar experiment, the apical polarized localization of de novo made SynA, used as a control of conventional secretion, is shown to be abolished in all three strains when RabE, AP‐1σ or ClaH is repressed. Scale bars: 5 μm. mCherry‐SynA PM/cytosolic intensity ratios are plotted with values being 0.52 ± 0.02 for wild type, 0.14 ± 0.02 for thiA p ‐rabE, 0.24 ± 0.02 for thiA p ‐ap1 σ and 0.16 ± 0.02 for thiA p ‐claH. The statistical evaluation was performed as in (A). There is a significant difference (****P < 0.0001) of mCherry‐SynA fluorescence in the apical PM membrane between the wild‐type and the three strains lacking the post‐Golgi proteins. Biological/technical replicates: 2/15 for each strain.
-
CCo‐localization analysis and relevant quantification of strains co‐expressing de novo made UapA‐GFP (120 min after initiation of transcription) with RabE‐mRFP, AP‐1‐mRFP or ClaH‐mRFP. Scale bars: 2 μm. Quantification by calculating Pearson's correlation coefficient (PCC) shows clear non‐co‐localization of UapA with all the Post‐Golgi markers tested, as confirmed by one sample t‐test (PCC = 0.31 ± 0.08, 0.29 ± 0.04 or 0.07 ± 0.01, with ****P < 0.0001, for RabE, AP‐1 or ClaH, respectively). Biological/technical replicates: 3/7 for each strain.
Source data are available online for this figure.
