Ovarian cancer (OC) has been called “the silent killer” with the 5-year survival rate reaching less than 40% that renders it the eight most common cause of cancer death in women and the second most deadly gynecologic cancer (1). Histologically OC can be divided in epithelial, germ cell and stromal subtypes. Among these, epithelial cancers are the most common and the most lethal (2). Epithelial OC includes serous (approximately 75% of all EOCs), endometrioid, clear cell and mucinous carcinomas. Among these subtypes, high-grade serous carcinoma (HGSOC) is the most prevalent histologic subtype of OC and, initially, extremely sensitive to chemotherapy (2,3). However, HGSOCs frequently relapse and become increasingly resistant to chemotherapy that results in treatment failure and patients’ death likely due to the selection of platinum resistant clones (4). Consequently, understanding the mechanisms underlying OC evolution and the onset of platinum resistance is an urgent unmet need in the management of OC patients that could help in establishing new targeted treatments or cutting-edge therapeutic approaches. In this context unveiling new hub signaling pathways necessary for EOC cells survival and spreading is a worth and recommended research effort that might unveil new therapeutic targets or envisage the repurposing of drugs originally used to treat different pathological conditions (5,6).
We have read with interest the recent report of Dahl and colleagues describing a previously undisclosed role for the isocitrate dehydrogenase 1 (IDH1) protein in OC. The authors propose that in HGSOC cells might represent a new therapeutic target to affect both metabolism and epigenetic regulation of HGSOC cells eventually leading to cancer cells senescence (7). Isocitrate dehydrogenases (IDHs) are crucial enzymes that coordinate different cellular processes, such as metabolism, epigenetic modifications, oxidative stress regulation and DNA repair (8), catalyzing the oxidative decarboxylation of isocitrate and producing α-ketoglutarate (αKG) (9). In humans, IDH family comprises three members: IDH1, IDH2, and IDH3. IDH3 is a NAD+-dependent enzyme that generates irreversibly αKG and NADH within the tricarboxylic acid (TCA) cycle. Conversely, IDH1 and IDH2 are homodimeric enzymes with a high degree of similarity, structurally and functionally distinct from the heterotrimeric IDH3. They are NADP+-dependent proteins that reversibly catalyze, in a redox reaction, the conversion of isocitrate to αKG (8,9). In the reverse reductive carboxylation reaction, citrate and acetyl-CoA are produced from isocitrate by reducing αKG. This activity is critical for the tumor cells, where anabolic process is maximized, to preserving lipids and cholesterol biosynthesis to sustain tumor cell proliferation (10). In addition, αKG promotes the activity of several αKG-dependent dioxygenases, like the Jumonji-domain containing histone-lysine demethylases (JHDMs) supporting a role of IDH1/2 in the regulation of epigenetic modifications, such as histone demethylation (11). It is well known that specific missense mutations in IDH1 and IDH2 have been identified in several tumor types, including grade II/III gliomas and secondary glioblastomas (GBM), chondrosarcomas, acute myeloid leukemias (AML), and less frequently in other types of cancers, like prostate cancers and melanomas (9). These mutations are usually somatic point mutations that bundle at the active site of the proteins driving to the alteration of crucial arginine residues (in IDH1, mutations mostly appear at R132, while for IDH2 at residues R172 and R140) critical for isocitrate binding and conferring to IDH1 and IDH2 a neomorphic activity able to convert αKG to the onco-metabolite D-2-hydroxyglutarate (D-2HG) (8,9). The main consequence is an abnormal cellular accumulation of the onco-metabolite mostly resulting in the inhibition of αKG-dependent dioxygenases that regulate epigenetic modifications, with a critical impact on gene expression profile and cancer cell proliferation (8,9). Moreover, the presence of IDH1/2 alterations impinge other different cellular pathways including metabolism, DNA damage repair and response to oxidative stress (8,9).
While oncogenic mutations in IDH1/2 genes have been extensively characterized in cancers, the implications of alterations in the expression of these enzymes and their possible therapeutic applications need more deeply investigations. IDH1, for example, is overexpressed in non-small cell lung carcinoma (NSCLC) and in adenocarcinoma and squamous cell carcinomas where it correlated with poor Overall Survival (OS), in approximately 65% of primary GBM and in several hematological malignancies (9).
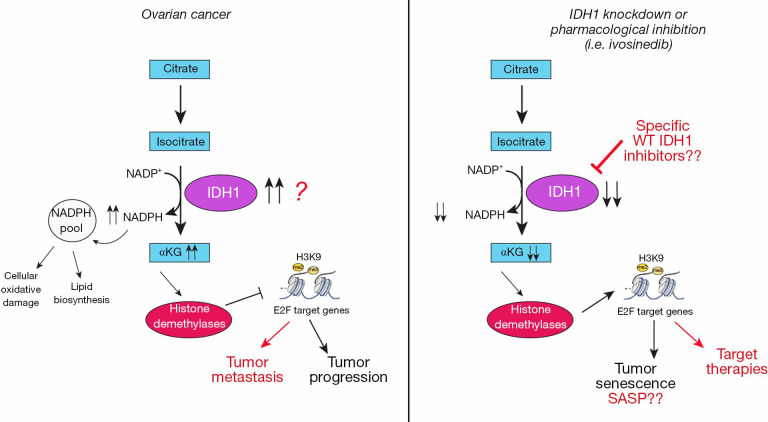
In this scenario, Dahl and colleagues highlighted the role of wild-type IDH1 overexpression in the regulation of metabolism and epigenetic modification in the context of OC. Starting from the study of the differences in the metabolic processes between normal fallopian tube and OC cells, the researchers observed a significant and consistent up regulation in the TCA cycle (tricarboxylic acid cycle, also know as Krebs cycle) in OC cells. Then the analysis of the expression of 27 enzymes implicated in TCA cycle regulation showed that IDH1 was the most significantly overexpressed in OC cells (both in adherent and in spheroids conditions). Looking at the expression of IDH1 in HGSOC patients’ samples, the authors also showed that IDH1 mRNA overexpression correlated with worse progression-free survival. Moreover, they demonstrated that OC cells privileged to use glucose for the oxidative decarboxylation reaction while the normal cells favored converting glucose to lactate, using aerobic glycolysis, to produce energy. These observations suggest that targeting glycolysis activity could be ineffective in treating OC cells, or rather, this approach can be harmful to the survival of normal cells. To go in deeper detail, the researchers found that the silencing or the pharmacological inactivation of IDH1 in HGSOC cells reduced BrdU incorporation, colony formation and expression of cell cycle progression markers, supporting the involvement of IDH1 in promoting cancer cell proliferation. Interestingly, the suppression of IDH1 in HGSOC cells induced senescence rather than cell death, as indicated by increased SA-α-Gal activity, decreased lamin B1 expression and increased PML bodies. Senescence is a cellular state of stable growth arrest induced by a wide range of intrinsic and extrinsic insults, like oncogenic activation, oxidative and genotoxic stress, shortened telomeres and chemotherapy (6). In this context, epigenetic modifications (i.e., histone methylations) modify the expression of genes involved in cell cycle progression and in the senescence-associated secretory phenotype (SASP) affecting, in turn, both the induction and suppression of senescence. For instance, one hallmark of the senescent cells is the increased expression of repressive histone marks, like the di- and tri-methylation marks of H3K9 at E2F target genes (12). As mentioned above, αKG is a required co-substrate for JHDMs histone demethylase, which remove the mono-, di-, and tri- methyl groups from the lysine residues of multiple histones, regulating chromatin state and, in turn, affecting different cellular processes (13). Several studies have shown a role for JmjC demethylases in altering the epigenome of senescent cells and, accordingly, Dahl and colleagues found an increased H3K9me2 occupancy at well-established E2F target genes as a consequence of αKG depletion upon IDH1 knock-down in OC cells. These results suggest that the impairment of IDH1 induce senescence by enhancing histone methylation at E2F target genes and repressing their expression. Interestingly, the authors found that SASP gene expression was not affected after IDH1 silencing. In the context of cancer research this is a very relevant observation since it is known that SASP can have both tumor suppressive and oncogenic activities (14). The data emerging from the Dahl group’s work support the idea that targeting IDH1 may promote a sustained block of cell cycle progression without the harmful side effects of the SASP, suggesting that, in the context of OC, senescence induction may overall be tumor suppressing (Figure 1).
Figure 1.
Schematic representation depicting the possible roles of IDH1 overexpression in Ovarian Cancer. IDH1 converts isocitrate to αKG and CO2, with concomitant production of NADPH from NADP+. In ovarian cancer cells (left panel), wild-type IDH1 up regulation increased TCA cycle metabolism and determines the increased amount of αKG and NADPH production which in turn provide high levels of reducing equivalents to sustain lipid biosynthesis and redox homeostasis and activate the αKG-dependent dioxygenases, histone demethylases. JHDM enzymes decrease histone H3K9 methylation on E2F target genes resulting in the activation of transcription of gene involved in cancer cell proliferation and tumor progression. IDH1 knockdown or its pharmacological inactivation (right panel) increased the repressive histone H3K9 methylation at multiple E2F target gene loci leading to ovarian cancer cell senescence. These data suggest that impairment of IDH1 may act as a new therapeutic target to affect both the metabolism and epigenetics of ovarian cancer cells and provide the rationale for the preclinical assessment of specific wild-type IDH1 inhibitors as anti-cancer agents (alone or in combination with other agents). Critical open questions in this pathway are marked with a question mark and include: the understanding of the mechanisms driving the overexpression of IDH1 in ovarian cancer cells; whether this upregulation also affects the ovarian cancer spreading and metastasis formation; if IDH1 inhibition cooperates with treatments currently available for ovarian cancer patients; if IDH1 WT inhibitors could be generated and used in vivo and why IDH1 inhibition does not lead to SASP production.
Overall we believe that the interesting work by Dahl and collaborators introduced the new concept that IDH1 wild type protein could act as a “druggable” oncogene in OC. This concept might prompt in the future the development of new specific molecules that better inhibits the wild type protein than the mutated one, as is for the available IDH1/2 inhibitors.
Alternatively, it would be interesting to verify if the inhibitors of H3K9 methyl-transferase (i.e., G9a and GLP) could be used to alter H3K9 methylation in the context of high IDH1 expression. Indeed several reports suggest that G9a might act as oncogene promoting metastasis and driving resistance to therapies in OC (15-17). At this regard, it is interesting to note that a specific G9a/GLP inhibitor can be safely used in preclinical models suggesting that this strategy could have future clinical applications also in cancer research (18).
Several stimulating questions that still require appropriate answers are also raised by the work of Dahl and colleagues (Figure 1). First it would be interesting to understand which are the OC types expressing high levels of IDH1 and if they belong to a specific subtype among the ones identified by gene expression profile studies (19,20). Also, it would be important to define if IDH1 overexpression is linked to gene amplification or to altered transcription. On a functional point of view it would be important to clarify why senescence associated to IDH1 knock-down in OC cell is not accompanied by the acquisition of a SASP phenotype. Also, it is unclear if IDH1 is associated to peritoneal dissemination and to the ability of OC cells to grow as spheroids in vivo. Finally, it would be important to clarify if IDH1 over expression has any role in the response to therapies currently used to treat OC patients including chemotherapy (e.g., platinum compounds and taxanes) and targeted therapies like PARP inhibitors and anti-angiogenic compounds. For instance, it is known that αKG alone is sufficient to suppress hypoxia-induced HIF1α expression, which could eventually result in altered tumor neoangiogenesis. Whether IDH1 expression could interfere with the activity of anti-angiogenic therapies in OC is still to be investigated.
Despite these still unanswered questions the work by Dahl et al. has the merit to open a new field of investigation in OC that might, in the future, provide new opportunities for a better classification of the disease and/or new therapeutic opportunities.
Of course many other genes are currently investigating as potential therapeutic targets in HGSOC that might merit being comment in more depth. However, this topic goes behind the scope of the present commentary and we hope that in the future the argument will be more extensively covered in a dedicated work.
Supplementary
The article’s supplementary files as
Acknowledgments
We like to thank all members of the SCICC lab for fruitful scientific discussion.
Funding: The study was supported by grants from Regione Autonoma Friuli Venezia Giulia (TICHEP and RIFT grants) and 5x1000 CRO to GB; grants from Ministero della Salute (RF-2016-02361040 to GB), (GR-2016-02361041 to MS).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Provenance and Peer Review: This article was commissioned by the editorial office, Annals of Translational Medicine. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.02.62). The authors have no conflicts of interest to declare.
References
- 1.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayson GC, Kohn EC, Kitchener HC, et al. Ovarian cancer. Lancet 2014;384:1376-88. 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 3.Matulonis UA, Sood AK, Fallowfield L, et al. Ovarian cancer. Nat Rev Dis Primers 2016;2:16061. 10.1038/nrdp.2016.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooke SL, Brenton JD. Evolution of platinum resistance in high-grade serous ovarian cancer. Lancet Oncol 2011;12:1169-74. 10.1016/S1470-2045(11)70123-1 [DOI] [PubMed] [Google Scholar]
- 5.Dall’Acqua A, Sonego M, Pellizzari I, et al. CDK6 protects epithelial ovarian cancer from platinum-induced death via FOXO3 regulation. EMBO Mol Med 2017;9:1415-33. 10.15252/emmm.201607012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonego M, Pellarin I, Costa A, et al. USP1 links platinum resistance to cancer cell dissemination by regulating Snail stability. Sci Adv 2019;5:eaav3235. [DOI] [PMC free article] [PubMed]
- 7.Dahl ES, Buj R, Leon KE, et al. Targeting IDH1 as a Prosenescent Therapy in High-grade Serous Ovarian Cancer. Mol Cancer Res 2019;17:1710-20. 10.1158/1541-7786.MCR-18-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molenaar RJ, Maciejewski JP, Wilmink JW, et al. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene 2018;37:1949-60. 10.1038/s41388-017-0077-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergaggio E, Piva R. Wild-Type IDH Enzymes as Actionable Targets for Cancer Therapy. Cancers (Basel) 2019. doi: . 10.3390/cancers11040563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011;481:380-4. 10.1038/nature10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006;7:715-27. 10.1038/nrg1945 [DOI] [PubMed] [Google Scholar]
- 12.Narita M, Nũnez S, Heard E, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003;113:703-16. 10.1016/S0092-8674(03)00401-X [DOI] [PubMed] [Google Scholar]
- 13.Cloos PA, Christensen J, Agger K, et al. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 2008;22:1115-40. 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao SG, Jackson JG. SASP: Tumor Suppressor or Promoter? Yes! Trends Cancer 2016;2:676-87. 10.1016/j.trecan.2016.10.001 [DOI] [PubMed] [Google Scholar]
- 15.Hua KT, Wang MY, Chen MW, et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer 2014;13:189. 10.1186/1476-4598-13-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J, Shin SH, Yoon H, et al. FIH Is an Oxygen Sensor in Ovarian Cancer for G9a/GLP-Driven Epigenetic Regulation of Metastasis-Related Genes. Cancer Res 2018;78:1184-99. 10.1158/0008-5472.CAN-17-2506 [DOI] [PubMed] [Google Scholar]
- 17.Watson ZL, Yamamoto TM, McMellen A, et al. Histone methyltransferases EHMT1 and EHMT2 (GLP/G9A) maintain PARP inhibitor resistance in high-grade serous ovarian carcinoma. Clin Epigenetics 2019;11:165. 10.1186/s13148-019-0758-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, Lee HM, Xiong Y, et al. Targeting the histone methyltransferase G9a activates imprinted genes and improves survival of a mouse model of Prader-Willi syndrome. Nat Med 2017;23:213-22. 10.1038/nm.4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 2008;14:5198-208. 10.1158/1078-0432.CCR-08-0196 [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as