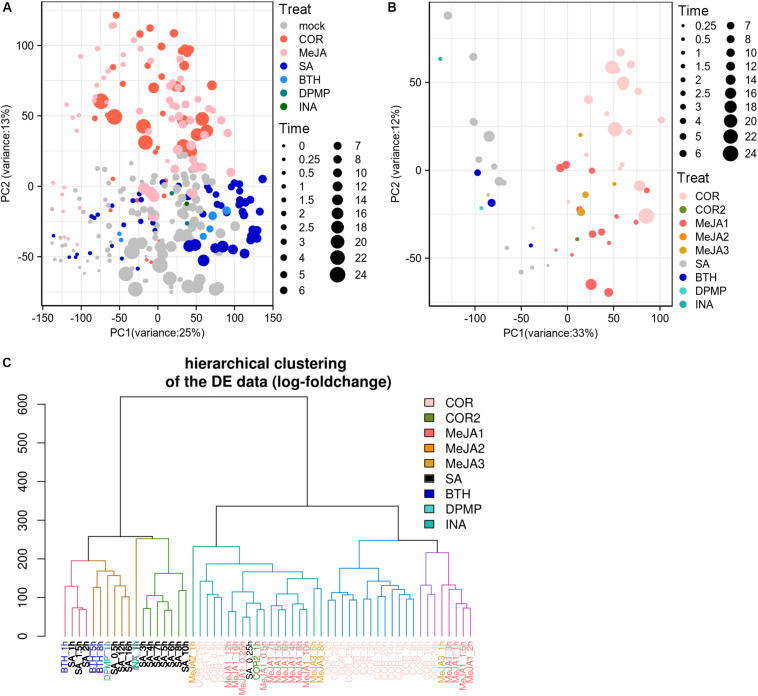
FIGURE 1.
JA/SA analogs were discriminated as two classes based on global differential gene expression matrix. (A,B) PCA plots showed the first 2 principal component diagram based on global gene expression matrix (after removing bath effects) and differential gene expression matrix. (C) HCA dendrogram profiled classification of global gene differential expression. Symbol sizes increased with time (hours) after the application of JA/SA analogs. The MeJA1-3 and COR/2 represented datasets from different laboratories. The RNA-Seq datasets: SRP041507 (COR); PRJNA224133 (SA/MeJA1); PRJNA354369 (BTH/MeJA2); PRJNA318266 (MeJA3); PRJNA270886 (COR2); PRJNA303108 (DPMP); PRJNA394842 (INA).