Abstract
Vibrio parahaemolyticus is the leading cause of seafood-borne bacterial poisoning in China and is a threat to human health worldwide. The aim of this study was to assess the antibiotic resistance profiles and distribution of heavy metal resistance of V. parahaemolyticus isolates from Penaeus vannamei from freshwater farms, seawater farms, and their corresponding markets in Zhejiang, China and to assess the relationship between multidrug resistance (MDR) and multi-heavy metal resistance (MHMR). Of the 360 P. vannamei samples that we tested, 90 (25.00%) were V. parahaemolyticus positive, but the occurrence of pathogenic isolates carrying the toxin genes tdh (4.44%) and trh (3.33%) was low. None of the tested isolates harbored both the tdh and trh genes. However, antibiotic resistance profiles varied among different sampling locations, levels of resistance to the antibiotics ampicillin (76.67%) and streptomycin (74.44%) were high overall, and MDR isolates were common (40.00% of all isolates). Heavy metal resistance patterns were similar among the different sampling locations. Overall, the majority of V. parahaemolyticus isolates displayed tolerance to Cd2+ (60.00%), and fewer were resistant to Cu2+ (40.00%), Zn2+ (38.89%), Ni2+ (24.44%), Cr3+ (14.44%), and Co2+ (8.89%). In addition, 34.44% (31/90) of isolates tested in this study were found to be MHMR. Using Pearson’s correlation analysis, MDR and MHMR were found to be positively correlated (P = 0.004; R = 0.759). The 18 V. parahaemolyticus isolates that were both MDR and MHMR represented 18 sequence types, of which 12 were novel to the PubMLST database, and displayed a high level of genetic diversity, suggesting that dissemination may be affected by mobile genetic elements via horizontal gene transfer. However, a low percentage of class 1 integrons without gene cassettes and no class 2 or 3 integrons were detected in the 18 MDR and MHMR isolates or in the 90 V. parahaemolyticus isolates overall. Thus, we suggest that future research focus on elucidating the mechanisms that lead to a high prevalence of resistance determinants in V. parahaemolyticus. The results of this study provide data that will support aquatic animal health management and food safety risk assessments in the aquaculture industry.
Keywords: antibiotic resistance, heavy metal resistance, integrons, Penaeus vannamei, sequence type diversity, Vibrio parahaemolyticus, virulence genes
Introduction
Vibrio parahaemolyticus, which was first identified in 1950 in Osaka, Japan, is a gram-negative, halophilic, mesophilic, aerobic bacterium that is found naturally in warm marine and estuarine habitats and causes outbreaks worldwide (Baker-Austin et al., 2008; Xie et al., 2017; Yang et al., 2017; He et al., 2019). Some V. parahaemolyticus isolates are pathogenic to humans and are responsible for many seafood-related human illnesses, such as gastrointestinal illnesses, diarrheal diseases, wound infections, and even septicemia (Devi et al., 2009; Hu and Chen, 2016; Yu et al., 2016). In addition, according to the official surveillance statistics of the national foodborne disease surveillance system in China, V. parahaemolyticus is one of the leading causes of foodborne bacterial poisoning in China (Jiang et al., 2019). The virulence of V. parahaemolyticus is attributed mainly to the presence of two genes: the tdh gene, which encodes thermostable direct hemolysin (TDH), and the trh gene, which encodes thermostable direct-related hemolysin (TRH) (Mok et al., 2019). TDH is a pore-forming, heat-stable protein that remains intact even when heated to 100°C for 10 min. Unlike the thermostable TDH, TRH is heat labile and can be inactivated by heating at 60°C for 10 min (Tan et al., 2017).
The shrimp aquaculture industry accounts for 15% of all internationally traded seafood products (FAO, 2019)1. Penaeus vannamei, also known as the Pacific whiteleg shrimp, is the most popular shrimp in the world. The production of P. vannamei exceeds 70% of total global shrimp production (Peng et al., 2019). It is also one of the most popular shrimp species in China, and China has been the world’s largest producer of P. vannamei since 2001 (FAO, 2019)1. P. vannamei is suitable for aquaculture owing to its fast growth, tolerance to a wide range of water salinity conditions and low temperatures, low dietary protein requirements, and high survival rates (Ghosh, 2018; Xu et al., 2018; Cheng et al., 2019). However, the increasing industrialization of large-scale intensive aquaculture systems and fast-growing shrimp culture models has led to poor hygienic conditions (Yu et al., 2016). As a consequence, the incidence of bacterial infection outbreaks is increasing (He et al., 2016; Yu et al., 2016). Shrimp represents an important reservoir of V. parahaemolyticus, especially in fresh and refrigerated stock (He et al., 2019). The distribution of V. parahaemolyticus in shrimps from China has previously been reported to be 22–55%, depending on the season, water salinity, and geographic location (Xu et al., 2016; He et al., 2019). The prevalence of V. parahaemolyticus is also high in other Asian countries; for example, the rate reported at shrimp farms in India is 35–53% (Silvester et al., 2015; Ananda Raja et al., 2017), and in Malaysia the rate was reported to be 57.8% in retail shrimps (Letchumanan et al., 2015). The use of florfenicol, thiamphenicol, enrofloxacin, flumequine, neomycin, doxycycline, ciprofloxacin, and certain sulfonamides is permitted in the aquaculture industry in China2. However, the inappropriate use of antibiotics in aquaculture has contributed to the development of antimicrobial resistant bacteria, imposed serious problems on aquatic ecosystems, and represents a potential threat to human health (He et al., 2016). It has been reported that V. parahaemolyticus isolates from seafood and various environments are resistant to a variety of antibiotics, including ampicillin, aminoglycosides, ciprofloxacin, chloramphenicol, and others (Xie et al., 2016; Lopatek et al., 2018). Thus, it is important to monitor variations in the antibiotic resistance profiles of V. parahaemolyticus isolates, as they may reveal changes in the sensitivity of the bacteria to antibiotics, particularly first-line treatments of seafood or human infections (Lopatek et al., 2018). In addition, industrial pollution caused by increased industrialization has become one of the most challenging issues facing developing countries (Kang et al., 2018). Among the many industrial pollutants, heavy metals are frequently detected in marine animals and in various environments, such as agricultural soil and rivers (Ansari et al., 2008; Malik and Aleem, 2011). Heavy metals have also been suggested to enhance selection for antibiotic resistance in the environment and vice versa through co- or cross-resistance or coregulation of resistance pathways (Matyar et al., 2008).
The mechanism of coselection is highly favored when diverse resistance genes are located on the same mobile genetic elements (MGEs; Chapman, 2003). Of the various MGEs, class 1 integrons are thought to be strictly correlated to coselection mechanisms, as they are frequently associated with gene cassettes (GCs) in which both antibiotic resistance genes and heavy metal resistance genes are present (Di Cesare et al., 2016). It has been reported that the clinical version of the class 1 integron-integrase gene (intI1) has unique advantages as a universal marker of the selective pressures imposed by anthropogenic pollution (Gillings et al., 2015).
Multilocus sequence typing (MLST) is a tool for molecular epidemiology and population genetic studies of bacterial strains that provides consistent typing results of bacterial isolates in different laboratories (Jiang et al., 2019). González-Escalona et al. (2008) developed the first successful MLST protocol for the detection of genetically diverse V. parahaemolyticus isolates based on the sequences of internal fragments of seven housekeeping genes (recA, gyrB, dnaE, dtdS, pntA, pyrC, and tnaA). Subsequently, many researchers utilized this method to determine the genetic relatedness of global and geographically restricted V. parahaemolyticus isolates and to demonstrate the evolution and epidemiology of the bacteria, as this method offers high repeatability (Theethakaew et al., 2013; Urmersbach et al., 2014; Han et al., 2015; Xie et al., 2016; Lopatek et al., 2018; Jiang et al., 2019). MLST also allows for the detection of slowly progressing sequence changes in the V. parahaemolyticus genome and may be used to monitor the spread of antibiotic and heavy metal resistance (Lopatek et al., 2018).
Zhejiang, a province in the southeastern coastal region of China, is an important area for P. vannamei aquaculture. In cities in Zhejiang that are not located next to the sea, freshwater with low salinity is used for P. vannamei aquaculture, whereas seawater is used in coastal cities (Cheng et al., 2019). In this study, we analyzed V. parahaemolyticus isolates from P. vannamei from freshwater and seawater farms and their corresponding markets in Zhejiang, China to assess their virulence genes and antibiotic and heavy metal resistance profiles. Subsequently, we determined the relationship between multidrug resistance (MDR) and multi-heavy metal resistance (MHMR) of V. parahaemolyticus isolates as well as the clonal relatedness of those isolates. Finally, we assessed the role of integrons in the transmission of antibiotic and heavy metal resistance. This information will contribute to the monitoring of the prevalence of antibiotic and heavy metal resistance of V. parahaemolyticus isolated from P. vannamei and provide insight into the appropriate use of antibiotics and the establishment of risk assessment and health management protocols for aquaculture and seafood consumption.
Materials and Methods
P. vannamei Sampling
A total of 360 P. vannamei samples were collected from a freshwater farm (farm A, n = 90), a seawater farm (farm B, n = 90), a market where P. vannamei cultured at farm A is sold (market A, n = 90) and a market where P. vannamei cultured at farm B is sold (market B, n = 90) in Zhejiang Province, China between 2017 and 2019. Samples were collected from each site during the period from July to September (summer) each year. Each fresh P. vannamei sample was placed in a sealed sterile plastic bag (Hope Bio-Technology Co., QingDao, China), transported to the laboratory in a cold box below 4°C, and immediately processed on the day of sampling. The pH value, temperature, and water salinity of each sampling site were recorded using a YSI Professional Plus Instrument (YSI Inc., Yellow Springs, OH, United States).
Isolation and Identification of V. parahaemolyticus
Each fresh P. vannamei sample (10–20 g) was mixed with alkaline peptone water (APW; Hope Bio-Technology Co.) containing 3% NaCl (1:1, w/v) in a sealed sterile plastic bag, and the mixture was homogenized for 2 min in a homogenizer (Scientz, Ningbo, China). Homogenates were incubated at 37°C with shaking at 200 rpm for 16–18 h. After incubation, the enriched mixture was streaked onto thiosulfate–citrate–bile salts–sucrose (TCBS, Hope Bio-Technology Co.) agar plates and incubated at 37°C for 16–18 h. Presumptive colonies (green or bluish-green colonies, 2–3 mm in diameter) were selected from each plate and streaked onto chromogenic Vibrio agar plates (CHROMagar Microbiology, Paris, France). Because the microbial cells of the aforementioned homogenates were enriched with many of the same colonies, we selected one potential colony (a purple colony, 2–3 mm in diameter) from each sample on the chromogenic Vibrio agar plates that represented the characteristics of V. parahaemolyticus. The potential colonies were cultured in APW containing 3% NaCl at 37°C for 24 h with shaking at 200 rpm and stored in 20% sterile glycerol at −80°C until further analysis.
Molecular Identification and Virulence Gene Detection of V. parahaemolyticus Isolates
Polymerase chain reaction (PCR) was used to detect the highly conserved species-specific gene toxR and the virulence genes tdh and trh in all V. parahaemolyticus isolates (Law et al., 2017). Genomic DNA was extracted using a bacterial DNA extraction kit (Sangon, Shanghai, China) according to the manufacturer’s instructions. The primers used to detect toxR, tdh and trh are shown in Table 1. Each PCR amplification reaction was performed in a 25 μL mixture containing 250 ng of DNA as the template, 400 nM each primer, 200 mM each deoxynucleotide triphosphate (dNTP), 10 × PCR buffer, and 5 U of Ex-Taq DNA polymerase (Takara-Bio, Beijing, China). PCR amplification was initiated by incubating the reaction mixture at 94°C for 1 min, followed by 30 cycles at 98°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and a final extension at 72°C for 10 min (Jiang et al., 2019). PCR products (5 μL) were mixed with 1 μL of 6 × loading buffer dye and analyzed by electrophoresis on a 1.2% agarose gel containing GoldView (Sangon, Shanghai, China). V. parahaemolyticus isolates ATCC33847 (tdh+ and trh–) and ATCC17802 (tdh– and trh+) were used as positive control isolates, and distilled water was used as the negative control.
TABLE 1.
Polymerase chain reaction primers used in this study.
| Target | Primer | Sequence (5′–3′) | Amplicon length (bp) | References |
| toxR | toxRF | TGTTTGGCGTGAGCAAGGTT | 340 | Primers used in our lab |
| toxRR | ATTCACAGCAGAAGCCACAG | |||
| Trh | trhF | TTGGCTTCGATATTTTCAGTATCT | 500 | Silva et al., 2018 |
| trhR | CATAACAAACATATGCCCATTTCCG | |||
| Tdh | tdhF | GGTACTAAATGGTTGACATC | 251 | Jiang et al., 2019 |
| tdhR | CCACTACCACTCTCATATGC | |||
| intI1 | intI1F | GGCTTCGTGATGCCTGCTT | 146 | Luo et al., 2010 |
| intI1R | CATTCCTGGCCGTGGTTCT | |||
| intI2 | intI2L | CACGGATATGCGACAAAAAGGT | 789 | Odumosu et al., 2013 |
| intI2R | GTAGCAAACGAGTGACGAAATG | |||
| intI3 | intI3L | GCCTCCGGCAGCGACTTTCAG | 980 | Odumosu et al., 2013 |
| intI3R | ACGGATCTGCCAAACCTGACT | |||
| Cassette arrays in class 1 integron | hep58 | TCATGGCTTGTTATGACTGT | Class 1 integron variable region | Odumosu et al., 2013 |
| hep59 | GTAGGGCTTATTATGCACGC |
Antibiotic Susceptibility Testing
The susceptibility of the confirmed V. parahaemolyticus isolates to 18 antibiotics was assessed using the disk diffusion method on Mueller–Hinton (MH) agar (Oxoid Ltd., Basingstoke, United Kingdom) in accordance with the guidelines of the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2017). The 18 antibiotic disks (Hangzhou Microbial Reagent Co., Hangzhou, China) belonged to 8 different classes and were as follows: quinolones (enrofloxacin, 10 μg; ciprofloxacin, 5 μg; norfloxacin, 10 μg), tetracyclines (tetracycline, 30 μg; doxycycline, 30 μg), aminoglycosides (streptomycin, 10 μg), β-lactams (ampicillin, 10 μg; cefazolin, 30 μg; cefamandole, 30 μg; ceftizoxime, 30 μg; cefepime, 30 μg; imipenem, 10 μg), chloramphenicols (chloramphenicol, 30 μg; florfenicol, 30 μg), sulfonamides (trimethoprim–sulfamethoxazole, 23.75 μg/1.25 μg; sulfisoxazole, 300 μg), polypeptides (polymyxin B, 300 μg), and furans (nitrofurantoin, 300 μg). Briefly, fresh cultures were inoculated into LB broth and incubated to a turbidity equivalent to a 0.5 McFarland standard. In a sterile environment, bacterial cultures were placed onto MH agar (Hope Bio-Technology Co.) plates, followed by antibiotic disks. The inoculated MH agar plates were incubated at 37°C for 24 h, and the size of the clear zone of inhibition was used to classify isolates as susceptible, intermediate, or resistant according to Clinical and Laboratory Standards Institute [CLSI] (2017) guidelines. Escherichia coli ATCC 25922 was used as a control (Jiang et al., 2019). Isolates resistant to three or more classes of antibiotics were classified as MDR (Cheng et al., 2019).
Determination of Heavy Metal Resistance of V. parahaemolyticus Isolates
To date, no standard method is available to measure bacterial susceptibility to heavy metals (He et al., 2016). According to the method described by Malik and Aleem (2011) with some modifications, the minimum inhibitory concentration (MIC) of heavy metals was determined for each V. parahaemolyticus isolate using MH agar containing Zn2+, Cu2+, Cd2+, Ni2+, Co2+, and Cr3+ in varying concentrations (100–3200 μg/mL). Stock solutions of metal salts were prepared in sterilized deionized water and added to MH agar at various concentrations, followed by spot inoculation with approximately 3 × 106 cells. The plates were then incubated at 37°C for 18–24 h. The metals used were ZnCl2, CuSO4⋅5H2O, NiCl2, CdCl2⋅5H2O, CoCl2⋅6H2O, and CrCl3⋅6H2O (Shanghai Macklin Biochemical Co., Ltd., Shanghai, China). Isolates were considered resistant if their MIC values exceeded that of the C600 strain of E. coli K-12, which was used as a control (Matyar et al., 2008). Isolates resistant to three or more heavy metals were classified as MHMR.
MLST Analysis
The clonal relatedness of 18 V. parahaemolyticus isolates that were both MDR and MHMR were further analyzed by MLST analysis. Seven housekeeping genes (recA, gyrB, dnaE, dtdS, pntA, pyrC, and tnaA) were chosen as target genes according to the PubMLST website.3 PCR primers, amplification conditions, and sequencing methods are described on the PubMLST website. The sequencing results for each housekeeping gene were analyzed using PubMLST to assign sequence types (STs). If STs or alleles were found to differ from preexisting ones in the database, the strain information of the new STs or the new allelic profiles with forward and reverse trace files were submitted to the database curator to obtain a new serial number (Jiang et al., 2019).
Detection of Integron Classes and GCs
The presence of integrase genes intI1, intI2, and intI3 and GCs was confirmed in all 90 V. parahaemolyticus isolates using PCR with specific primers (Table 1). PCR amplification was performed in a 25 μL mixture as described in the foregoing, and reaction conditions included preincubation at 94°C for 1 min, followed by 35 cycles of denaturation at 98°C for 30 s, annealing at 60°C (intI1) or 55°C (intI2 and intI3) for 30 s, and elongation at 72°C for 30 s (intI1) or 1 min (intI2 and intI3); and a final extension at 72°C for 10 min (Fang et al., 2019).
Because the intI2 and intI3 genes were not detected in any of our isolates, the variable regions (VRs) of isolates that were positive for the intI1 gene were evaluated using PCR. Class 1 integron VRs were amplified using the primers hep58 and hep59 (Table 1) and the following cycling conditions: preincubation at 94°C for 1 min; followed by 35 cycles of denaturation at 98°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for 4 min; and a final extension at 72°C for 10 min (Fang et al., 2019).
Statistical Analysis
All experiments were performed in triplicate. Correlations were identified using Pearson’s correlation analysis. The degree of correlation was considered weak if the correlation coefficient (R) was < 0.4, moderate if R was between 0.4 and 0.6, and strong if R was ≥ 0.6. Differences were considered significant when P-values were < 0.05. All analyses were performed using SPSS version 20.0 (IBM Corp., Armonk, NY, United States).
Results
Physicochemical Properties of Each Sampling Site
Analytical data for various physicochemical parameters (pH value, temperature, and water salinity) were collected from the four different sampling sites. As shown in Table 2, at the four sampling sites temperature varied from 29.10 ± 0.03 to 32.50 ± 0.05°C, and pH ranged from 6.92 ± 0.03 to 8.00 ± 0.20, which is within the permissible range for shrimp culture. In addition, water salinity varied over a wide range, and was low at farm A (1.75 ± 0.03 ppt) and market A (1.43 ± 0.02 ppt), but was high at farm B (27.10 ± 0.09 ppt) and market B (23.70 ± 0.18 ppt).
TABLE 2.
Physicochemical parameters of the four sampling sites.
| Sources of samples | Temperature (°C) | pH | Salinity (ppt) |
| Farm A | 31.53 ± 0.05 | 8.00 ± 0.20 | 1.75 ± 0.03 |
| Farm B | 32.50 ± 0.05 | 7.52 ± 0.12 | 27.10 ± 0.09 |
| Market A | 29.10 ± 0.03 | 7.64 ± 0.01 | 1.43 ± 0.02 |
| Market B | 30.18 ± 0.09 | 6.92 ± 0.03 | 23.70 ± 0.18 |
Identification of V. parahaemolyticus Isolates and Detection of tdh and trh Genes
Based on the morphology of colonies on chromogenic Vibrio agar plates, 90 (25.00%) isolates from the 360 P. vannamei samples were selected for PCR detection. ToxR-PCR assays revealed positive amplification of the toxR gene, with a 340 bp amplicon band present in 100% of the presumptive V. parahaemolyticus isolates, including 3 (3.33%) of the 90 samples from farm A, 31 (34.44%) of the 90 samples from farm B, 20 (22.22%) of the 90 samples from market A, and 36 (40.00%) of the 90 samples from market B. Among these isolates, four from P. vannamei at farm B carried the tdh gene (4.44%), whereas three isolates from P. vannamei at market B carried the trh gene (3.33%). None of the tested isolates harbored both the tdh and trh genes.
Antibiotic Resistance Profiles of V. parahaemolyticus Isolates
The antibiotic resistance profiles of 90 examined V. parahaemolyticus isolates to 18 antibiotics from eight classes are shown in Table 3. The profiles varied among the four different sampling locations. Overall, V. parahaemolyticus isolates were most resistant to ampicillin and streptomycin, with resistance rates of 76.67% (69/90) and 74.44% (67/90), respectively. In addition, these isolates exhibited relatively high resistance rates for trimethoprim–sulfamethoxazole (64.44%, 58/90), tetracycline (57.78%, 52/90), chloramphenicol (57.78%, 52/90), florfenicol (53.33%, 48/90), enrofloxacin (47.78%, 43/90), sulfisoxazole (47.78%, 43/90), doxycycline (46.67%, 42/90), cefazolin (25.56%, 23/90), and cefamandole (18.89%, 17/90). Low levels of resistance were observed for ceftizoxime (4.44%, 4/90) and cefepime (3.33%, 3/90). None of our isolates demonstrated resistance to ciprofloxacin, norfloxacin, imipenem, polymyxin B, or nitrofurantoin. Furthermore, 40.00% (36/90) of the isolates were classified as MDR.
TABLE 3.
Prevalence of antimicrobial resistance in Vibrio parahaemolyticus isolates.
| Antimicrobials | Breakpoint (Clinical and Laboratory Standards Institute [CLSI], 2017) | Percentage of resistant isolates, % | |||||
| Resistant/Intermediate/Susceptible (mm) | Farm A (n = 3) | Farm B (n = 31) | Market A (n = 20) | Market B (n = 36) | Total (n = 90) | ||
| Quinolone | Enrofloxacin (10 μg) | ≤ 15/16–20/ ≥ 21 | 33.30 | 41.94 | 75.00 | 38.89 | 47.78 |
| Ciprofloxacin (5 μg) | ≤ 15/16–20/ ≥ 21 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Norfloxacin (10 μg) | ≤ 12/13–16/ ≥ 17 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Tetracycline | Tetracycline (30 μg) | ≤ 11/12–14/ ≥ 15 | 33.33 | 67.74 | 65.00 | 47.22 | 57.78 |
| Doxycycline (30 μg) | ≤ 10/11–13/ ≥ 14 | 33.30 | 67.74 | 35.00 | 36.11 | 46.67 | |
| Aminoglycoside | Streptomycin (10 μg) | ≤ 11/12–14/ ≥ 15 | 33.33 | 74.19 | 70.00 | 80.56 | 74.44 |
| β-Lactam | Ampicillin (10 μg) | ≤ 13/14–16/ ≥ 17 | 66.67 | 61.29 | 85.00 | 86.11 | 76.67 |
| Cefazolin (30 μg) | ≤ 19/20–22/ ≥ 21 | 33.30 | 19.35 | 35.00 | 25.00 | 25.56 | |
| Cefamandole (30 μg) | ≤ 14/15–17/ ≥ 18 | 0.00 | 25.81 | 15.00 | 16.67 | 18.89 | |
| Ceftizoxime (30 μg) | ≤ 21/22–24/ ≥ 25 | 0.00 | 6.45 | 5.00 | 2.78 | 4.44 | |
| Cefepime (30 μg) | ≤ 18/19–24/ ≥ 25 | 0.00 | 3.23 | 5.00 | 2.78 | 3.33 | |
| Imipenem (10 μg) | ≤ 13/14–15/ ≥ 16 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Chloramphenicol | Chloramphenicol (30 μg) | ≤ 12/13–17/ ≥ 18 | 33.33 | 61.29 | 45.00 | 63.89 | 57.78 |
| Florfenicol (30 μg) | ≤ 12/13–17/ ≥ 18 | 0.00 | 67.74 | 40.00 | 52.78 | 53.33 | |
| Sulfonamides | Trimethoprim–sulfamethoxazole (23.75/1.25 μg) | ≤ 10/11–15/ ≥ 16 | 66.67 | 83.87 | 50.00 | 55.55 | 64.44 |
| Sulfisoxazole (300 μg) | ≤ 12/13–16/ ≥ 17 | 33.33 | 54.84 | 50.00 | 41.67 | 47.78 | |
| Polypeptide | Polymyxin B (300 μg) | ≤ 8/9–11/ ≥ 12 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Furan | Nitrofurantoin (300 μg) | ≤ 14/15–16/ ≥ 17 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| MDR | 33.33 | 38.71 | 40.00 | 41.67 | 40.00 | ||
Tolerance of V. parahaemolyticus Isolates to Heavy Metals
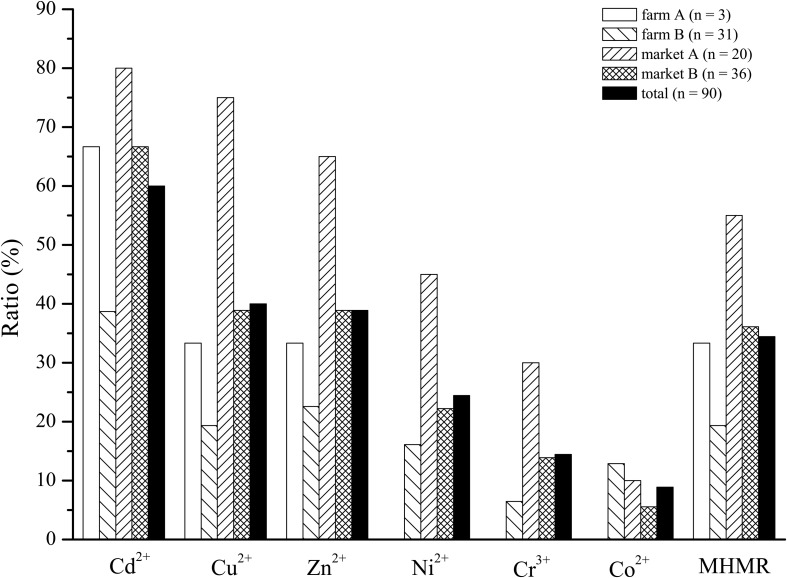
In this study, a total of 90 V. parahaemolyticus isolates were examined for their susceptibility to heavy metals. When compared to the E. coli K-12 C600 quality control strain, maximum MIC values of 3200 μg/mL for Cd2+, Cu2+, Zn2+, and Ni2+ and 1600 μg/mL for Cr3+ and Co2+ were observed (Table 4). Overall, the majority of V. parahaemolyticus isolates displayed tolerance to Cd2+ (60.00%), and fewer were resistant to Cu2+ (40.00%), Zn2+ (38.89%), Ni2+ (24.44%), Cr3+ (14.44%), and Co2+ (8.89%). In total, 34.44% (31/90) of isolates in this study were found to be resistant to three or more heavy metals and were classified as MHMR. In addition, as shown in Figure 1, V. parahaemolyticus isolates derived from different shrimp sources had similar heavy metal resistance profiles.
TABLE 4.
Incidence of heavy metal resistance among the V. parahaemolyticus isolates.
| Heavy metal | MIC (μg/mL) |
Resistance |
||||||
| 100 | 200 | 400 | 800 | 1600 | 3200 | n | (%) | |
| Cd2+ | 5 | 31a | 29 | 17 | 6 | 2 | 54 | 60.00 |
| Cu2+ | 6 | 9 | 10 | 29a | 21 | 15 | 36 | 40.00 |
| Zn2+ | 4 | 19 | 32a | 14 | 18 | 3 | 35 | 38.89 |
| Ni2+ | 10 | 15 | 43 a | 7 | 13 | 2 | 22 | 24.44 |
| Cr3+ | 0 | 1 | 21 | 55a | 13 | 0 | 13 | 14.44 |
| Co2+ | 2 | 80a | 4 | 3 | 1 | 0 | 8 | 8.89 |
| MHMR | 31 | 34.44 | ||||||
aMinimal inhibitory concentration of standard strain E. coli K-12.
FIGURE 1.
Incidence of heavy metal resistance of Vibrio parahaemolyticus isolates derived from four different sampling sites.
Relationship Between Pathogenicity, MDR, and MHMR
In our study, although none of the four isolates harboring the tdh gene or the three isolates harboring the trh gene were MDR or MHMR, our data are insufficient to conclude that there was a correlation between pathogenicity and MDR or between pathogenicity and MHMR. However, MDR and MHMR were found to be positively correlated using Pearson’s correlation analysis (P = 0.004; R = 0.759).
ST Diversity
MLST analysis revealed high molecular diversity among the 18 V. parahaemolyticus isolates in this study that were both MDR and MHMR (Table 5). A total of 18 different STs were identified, of which 12 (66.67%), namely 2217–2228, were newly identified (shown in bold in Table 5). The numbers of alleles observed for each MLST locus in our study were as follows: 14 dnaE, 14 gyrB, 11 recA, 14 dtdS, 10 pntA, 13 pyrC, and 12 tnaA. There were also seven novel loci: dnaE 401; gyrB 539, 540; dtdS 484, 485, 486; and pntA 271. All 12 novel STs were submitted to the international PubMLST/V. parahaemolyticus database3.
TABLE 5.
Allele profiles, sequence types (STs), virulence genes, integrons, antimicrobial resistance profiles, and heavy metal resistance profiles of 18 multidrug resistant and multi-heavy metal resistant Vibrio parahaemolyticus isolates.
| Strain | Sources | Allele profiles |
STs | Virulence genes |
Integrons | Antimicrobial resistance profiles | Heavy metal resistance profiles | |||||||
| dnaE | gyrB | recA | dtdS | pntA | pyrC | tnaA | tdh | trh | ||||||
| MA-3 | Market A | 2 | 113 | 28 | 94 | 26 | 83 | 23 | 2217 | - | - | - | ENR-TET-STR-AMP | Zn2+, Cu2+, Cd2+, Ni2+ |
| MA-8 | Market A | 93 | 224 | 75 | 139 | 117 | 223 | 124 | 919 | - | - | - | ENR-TET-STR-AMP | Zn2+, Cu2+, Cd2+, Ni2+ |
| MA-17 | Market A | 47 | 112 | 75 | 97 | 68 | 69 | 64 | 2221 | - | - | + | ENR-TET-STR-AMP-CHL-FLO | Zn2+, Cu2+, Ni2+ |
| FB-11 | Farm B | 76 | 88 | 31 | 13 | 53 | 45 | 13 | 165 | - | - | + | ENR-TET-DOX-AMP-CFZ-ZOX-FEP | Zn2+, Cu2+, Cd2+ |
| FB-15 | Farm B | 175 | 22 | 168 | 201 | 130 | 17 | 73 | 471 | - | - | - | ENR-STR-AMP-CFZ-ZOX | Zn2+, Cu2+, Ni2+, Cr2+ |
| FB-16 | Farm B | 3 | 104 | 144 | 126 | 28 | 226 | 159 | 2219 | - | - | - | TET-DOX-STR-AMP | Zn2+, Cu2+, Ni2+, Cr2+ |
| FB-21 | Farm B | 82 | 539 | 134 | 373 | 26 | 3 | 17 | 2223 | - | - | - | ENR-TET-AMP-CFZ-ZOX-CHL-SXT-SUL | Zn2+, Cu2+, Cd2+, Ni2+, Cr2+ |
| FB-23 | Farm B | 401 | 188 | 134 | 484 | 26 | 3 | 1 | 2224 | - | - | - | ENR-TET-STR-AMP-CFZ-MA-ZOX | Zn2+, Cu2+, Cd2+ |
| FB-31 | Farm B | 80 | 540 | 61 | 485 | 35 | 141 | 157 | 2225 | - | - | - | ENR-TET-AMP-SUL | Zn2+, Cu2+, Cr2+ |
| MB-2 | Market B | 5 | 71 | 144 | 486 | 26 | 11 | 107 | 2222 | - | - | - | ENR-TET-DOX-STR-CHL-FLO | Zn2+, Cu2+, Cd2+, Ni2+, Cr2+ |
| MB-7 | Market B | 51 | 4 | 25 | 76 | 271 | 173 | 33 | 2226 | - | - | - | ENR-TET-AMP-FEP-FLO-SUL | Zn2+, Cu2+, Ni2+, Co2+ |
| MB-11 | Market B | 36 | 304 | 188 | 218 | 28 | 168 | 37 | 2220 | - | - | - | ENR-STR-AMP-CHL-FLO | Zn2+, Cd2+, Co2+ |
| MB-14 | Market B | 175 | 112 | 168 | 201 | 68 | 69 | 64 | 2227 | - | - | - | ENR-TET-AMP-CFZ-FLO | Zn2+, Cu2+, Cd2+, Ni2+, Cr2+ |
| MB-17 | Market B | 175 | 22 | 25 | 201 | 130 | 17 | 73 | 2218 | - | - | - | ENR-TET-AMP-CHL-FLO | Zn2+, Cu2+, Cd2+, Ni2+ |
| MB-24 | Market B | 51 | 4 | 168 | 76 | 271 | 173 | 33 | 2228 | - | - | - | ENR-TET-STR-AMP-MA-ZOX-FEP | Zn2+, Cu2+, Cd2+, Ni2+ |
| MB-29 | Market B | 175 | 49 | 168 | 201 | 130 | 17 | 73 | 2174 | - | - | - | TET-STR-AMP-MA-ZOX-FEP-CHL-SUL | Zn2+, Cu2+, Ni2+, Co2+ |
| MB-32 | Market B | 153 | 191 | 70 | 19 | 6 | 8 | 1 | 1160 | - | - | - | ENR-TET-STR-AMP-ZOX-SUL | Zn2+, Cu2+, Cd2+, Co2+, Cr2+ |
| MB-34 | Market B | 98 | 49 | 45 | 49 | 4 | 50 | 23 | 1786 | - | - | - | TET-DOX-STR-AMP-CFZ-CHL-FLO | Zn2+, Cu2+, Cd2+, Ni2+, Co2+, Cr2+ |
Newly identified STs are shown in boldface type. AMP, ampicillin; CFZ, cefazolin; CHL, chloramphenicol; CIP, ciprofloxacin; DOX, doxycycline; ENR, enrofloxacin; FEP, cefepime; FLO, florfenicol; IPM, imipenem; MA, cefamandole; NFX, norfloxacin; NIT, nitrofurantoin; POL, polymyxin B; STR, streptomycin; SUL, sulfisoxazole; SXT, trimethoprim–sulfamethoxazole; TET, tetracycline; ZOX, ceftizoxime.
Integron Analysis
Integrons were examined in all 90 V. parahaemolyticus isolates. We found that eight isolates (8.89%) harbored class 1 integrons, but no class 2 or 3 integrons were detected. Of the 8 class 1 integron-positive isolates, 4 (11.11%) were recovered from 36 MDR isolates, 3 (9.68%) were recovered from 31 MHMR isolates, and 2 (11.11%) were recovered from 18 MDR and MHMR isolates. No isolates contained GCs. Correlation analysis showed that neither MDR (P = 0.550; R = 0.192) nor MHMR (P = 0.990; R = 0.004) was significantly correlated with the carrying of V. parahaemolyticus class 1 integrons.
Discussion
The occurrence of V. parahaemolyticus in aquatic samples has raised increasing concern worldwide, as this organism is one of the leading nationwide causes of food-derived bacterial poisoning in humans (Elmahdi et al., 2016; Zhao et al., 2020). In our study, only 3 V. parahaemolyticus isolates were detected from 90 samples of freshwater P. vannamei cultured at farm A, where the water salinity was as low as 1.75 ± 0.03 ppt. One possible reason for such a low occurrence of V. parahaemolyticus isolates is that, within a limited optimal temperature range, water salinity is a driver of V. parahaemolyticus levels (Zimmerman et al., 2007). As a halophilic bacterium, the survival of V. parahaemolyticus in freshwater ecosystems has been shown to be transient and dependent on the biological host (Nair et al., 2007). Freshwater cultured P. vannamei requires multiple gradients to slowly acclimate seedlings to reduced salinity (Peng et al., 2019), and during this process V. parahaemolyticus cannot be completely eliminated. However, 20 (22.22%) V. parahaemolyticus isolates were found at market A, where freshwater P. vannamei cultured at farm A is sold and the water salinity of market A is low, too. Previously, the presence of V. parahaemolyticus in freshwater samples from markets was attributed to cross-contamination due to mishandling at fishmongers’ stalls (Nair et al., 2007). Because the surrounding environment of markets is quite complex, some V. parahaemolyticus isolates may have been transmitted from the surrounding marine food in the market via human contact, water sources, or other animals. In addition, prevalence rates of V. parahaemolyticus were found to be 34.44% at seawater farm B and 40.00% at market B, where P. vannamei cultured at farm B is sold, which is consistent with previous studies that assessed marine products in China (Xu et al., 2016; Xie et al., 2017). In our study, water salinity varied over a wide range and was low at farm A and market A but was high at farm B and market B. On the one hand, P. vannamei is euryhaline and can tolerate salinity ranging from 1 to 50 ppt (Jaffer et al., 2020). On the other hand, in order to keep P. vannamei alive and fresh, water salinity at markets should be similar to that at the farm where the P. vannamei was cultured. Similarly, a suitable temperature and pH value should be maintained and an aerator used, as consumers at Asian markets prefer live shrimp to dead shrimp (Xu et al., 2019).
As previously described, the hemolysin markers tdh and trh play a significant role in the pathogenesis of human infections (Lopatek et al., 2018). In the present study, the tdh gene was detected at slightly higher levels, whereas the trh gene was detected at lower levels, than those reported in previous studies (Xie et al., 2017; Lopatek et al., 2018). However, other studies reported detecting no virulence genes in aquatic products (Han et al., 2007; Raghunath et al., 2008; Xu et al., 2016). The distribution of tdh- and trh-positive isolates may vary depending on the sample source, the detection technique, and the geographical origin (Lopatek et al., 2018). It has been reported that clinical isolates have higher rates of virulence genes than isolates from aquatic products, which may be due to environmental factors such as interactions with other hosts and the evolution of pathogens (Letchumanan et al., 2015; Xie et al., 2017). Additionally, tdh-positive isolates are more virulent than trh-positive isolates (Kang et al., 2018). In our study, none of the isolates from markets A and B were positive for tdh, which may represent a reduced risk for serious infections for consumers.
Antibiotic susceptibility testing revealed that V. parahaemolyticus isolates were most resistant to ampicillin and streptomycin, with resistance rates of 76.67 and 74.44%, respectively. These results are comparable to data obtained in other countries (Lopatek et al., 2018). It has been reported that the prevalence of ampicillin and streptomycin resistance is very high in V. parahaemolyticus from both clinics and aquatic products (Wong et al., 2012; Xie et al., 2017; Silva et al., 2018). This may be due to the extensive use of these antibiotics in aquaculture and antimicrobial residues in aquatic systems (Tan et al., 2017). In addition, compared with gram-positive species, gram-negative bacteria are intrinsically less permeable, which may allow them to resist certain antibiotics, as their outer membrane forms a permeability barrier (Blair et al., 2015).
Tetracycline, ciprofloxacin, chloramphenicol, trimethoprim–sulfamethoxazole, and cephalosporin are first-line drugs used in the clinical treatment of V. parahaemolyticus infections and were tested in the present study (Xie et al., 2016; Tan et al., 2017; Yang et al., 2017; Mok et al., 2019). Our findings showed that 57.78, 57.78, and 64.44% of isolates were resistant to tetracycline, chloramphenicol, and trimethoprim–sulfamethoxazole, respectively, which is much higher than rates reported by other studies in several countries and for several sample sources (Yano et al., 2011; Ottaviani et al., 2013; Tan et al., 2017). However, none of our isolates demonstrated resistance to ciprofloxacin, indicating that this antibiotic is still highly effective against V. parahaemolyticus and can continue to be recommended as a therapeutic drug. Additionally, V. parahaemolyticus isolates in the present study were highly resistant to first- and second-generation cephalosporins (cefazolin, 25.56%; cefamandole, 18.89%); however, fewer than 5% of isolates were resistant to third- and fourth-generation cephalosporins (ceftizoxime, 4.44%; cefepime, 3.33%). This suggests that first- and second-generation cephalosporins may have been misused in the past decades, leading to reduced susceptibility and lower efficiency in the treatment of V. parahaemolyticus (Yu et al., 2016). However, the accumulation of third- and fourth-generation cephalosporins in the environment and resulting drug resistance may occur more slowly, and the small number of cases of drug resistance reported in the present study indicates potential future risks. Moreover, high resistance to third-generation cephalosporins has previously been reported in V. parahaemolyticus from shrimp in Malaysia, another Asian country (Letchumanan et al., 2019a). In addition, extended-spectrum β-lactamases (ESBLs) confer resistance to a broad range of β-lactams, including third- and fourth-generation cephalosporins, which are commonly found in Enterobacteriaceae (Tetens et al., 2019). However, recently, some studies found that the prevalence of ESBL genes varied among new generation β-lactam-resistant Vibrio sp. isolates (Bush and Fisher, 2011; Dahanayake et al., 2019). Thus, potential ESBL-mediated cephalosporin resistance mechanisms of V. parahaemolyticus should be further researched.
Antibiotics are widely used in aquaculture to control bacterial infections and promote the growth of aquatic organisms. Some of the antibiotics tested in our study, including chloramphenicol, norfloxacin, and nitrofurantoin, have already been banned in food-producing animals in China4 ,5. In the present study, none of the V. parahaemolyticus isolates demonstrated resistance to norfloxacin or nitrofurantoin. Interestingly, we observed high rates of resistance to chloramphenicol (57.78% overall), which has been banned in food-producing animals since 2002. Moreover, a high rate of resistance (53.33% overall) to florfenicol, a fluorinated derivative of chloramphenicol that is widely used to treat aquatic infections in many countries including China, Brazil, and the United States (Yang et al., 2020), was also observed in our study. We found similar results in our previous study of E. coli isolates from P. vannamei (Cheng et al., 2019). Homology analysis revealed that the sequence of the florfenicol resistance gene floR had 55% homology with the sequence of the chloramphenicol resistance gene cmlA, both of which are efflux transporters belonging to the major facilitator superfamily (Kadlec et al., 2007; Fang et al., 2019). Thus, we hypothesize that high levels of resistance to chloramphenicol may be related to the use of florfenicol (Yassin et al., 2017). Additionally, although the use of doxycycline, enrofloxacin, and sulfisoxazole is permitted in the aquaculture industry in China6, some V. parahaemolyticus isolates in our study were resistant to these antibiotics.
Finally, polymyxin B and imipenem are two special antibiotics tested in our study. Polymyxins, including polymyxin B and polymyxin E, have broad-spectrum activity against gram-negative bacteria but are typically considered last-resort antibiotics to treat severe infections caused by MDR isolates (Liu et al., 2016). Imipenem belongs to the carbapenem class of β-lactams, has a very broad spectrum of activity, and acts mostly on gram-negative and gram-positive bacteria (Cheng et al., 2019). Fortunately, none of the V. parahaemolyticus isolates in our study exhibited resistance to these two special antibiotics. However, other studies have reported resistance of Vibrio sp. to polymyxin B and imipenem. Misuse of polymyxins and carbapenem may have a negative impact on the clinical treatment of Vibrio infections in the future (Devi et al., 2009; Lee et al., 2018).
MDR isolates were commonly observed in our study (40.00% overall), which is consistent with findings in previous studies (Xu et al., 2016; Yang et al., 2017). There are many possible reasons for the existence of MDR V. parahaemolyticus isolates, including the excessive use of antibiotics for prophylactic use, therapeutic use, or as antimicrobial growth promoters within the aquaculture industry (Xie et al., 2017). The widespread use of antibiotics in clinics, agriculture, and livestock production can result in antibiotic residues entering the environment (Tan et al., 2017), and the exchange of genetic resistance determinants between different environments can occur via direct or indirect contact or via MGEs (Fang et al., 2019). Therefore, non-antibiotic approaches are required to manage the occurrence of antibiotic resistance among Vibrio sp. in the environment (Lee et al., 2018). Phages are approved and recognized by US regulatory bodies as potential biocontrol agents to control and inhibit pathogens, including Vibrio sp. (Letchumanan et al., 2016, 2019b). Phages pose significant advantages, such as being environmentally friendly and easily discoverable in the environment and having greater host specificity and cost effectiveness than antibiotics (Letchumanan et al., 2016). In addition, some probiotics are possible substitutes for antibiotics against Vibrio sp. Tan et al. (2020) identified bioactive compounds with anti-Vibrio activity from Streptomyces sp. that will be of immense value for the future development of antibacterial agents.
It has been reported that, owing to its unique geographical environment and the rapid expansion of aquacultural, industrial, and agricultural activities, the Zhejiang nearshore area has already suffered heavy metal contamination (Liang et al., 2019). Moreover, heavy metal resistance has been observed in Vibrio sp. isolated from aquatic products and the environment in many neighboring provinces of Zhejiang (He et al., 2016, 2019; Hu and Chen, 2016). Some of the heavy metals tested in this study, such as Cu, Zn, Ni, Co, and Cr, are essential micronutrients for bacteria at low concentrations. However, concentrations of such metals above threshold levels, as well as the long-term presence of potentially toxic metals (e.g., Cd in our study), adversely affect the functioning and diversity of microbial communities, which may eventually result in some bacteria developing resistance to heavy metals via complex formation or sequestration of toxic metals, detoxification through reduction of intracellular ions, or extrusion of toxic ions by efflux systems (López-Maury et al., 2002; Matyar et al., 2008; Seiler and Berendonk, 2012; Etesami, 2018). In addition, V. parahaemolyticus isolates derived from different shrimp sources had similar heavy metal resistance profiles. The results indicated that the sample source did not appear to greatly impact the heavy metal resistance profiles of V. parahaemolyticus isolates, which was also reported in a previous study (He et al., 2016). One possible reason is that industrial pollution may influence the aquaculture environment, as pollutants (e.g., heavy metals) concentrated in the region have a relatively significant impact on certain species and microbes.
In our study, MDR and MHMR were found to be positively correlated. It has been reported that industrial pollutants such as heavy metals may enhance selection for antibiotic resistance and vice versa. One possible reason for this is that metal cations are common environmental stressors that perturb bacteria, activating metal-protective stress responses and growth states that also protect against and provide resistance to antibiotics (Hu and Chen, 2016; Poole, 2017). Additionally, coselection or the coexistence of certain antibiotic resistance genes and heavy metal resistance genes may be beneficial to bacteria for increasing fitness in various environments. Importantly, 18 V. parahaemolyticus isolates that were both MDR and MHMR were resistant to Zn2 + and 17 of them were resistant to Cu2 +, although their antibiotic and heavy metal resistance profiles were different. Moreover, resistance to ampicillin (94.44%, 17/18), tetracycline (88.89%, 16/18), enrofloxacin (83.33%, 15/18), and streptomycin (66.67%, 12/18) occurred frequently in the 18 isolates. There is a growing body of evidence that Cu/Zn drives antibiotic resistance in metal-exposed bacteria owing to the selection of genetic elements harboring both antibiotic and metal resistance genes and to the recruitment of antibiotic resistance mechanisms by metals. Moreover, Cu2+ and Zn2+ can bind to certain classes of antibiotics (e.g., the β-lactams, aminoglycosides, tetracyclines, and quinolones tested in this study) to form complexes and hinder antibiotic activity (Poole, 2017). In any case, increases in MHMR and MDR in aquatic products play a crucial role in the food chain and may pose important public health problems (Matyar et al., 2008).
Similar to many previous studies, the STs of the V. parahaemolyticus isolates in our study were diverse, and most isolates represented novel STs, indicating a high degree of genomic diversity and suggesting that dissemination of MDR and MHMR genes of V. parahaemolyticus isolates from P. vannamei samples may be effected by MGEs via horizontal gene transfer (HGT) (Xie et al., 2016; Lopatek et al., 2018; Yang et al., 2017; Jiang et al., 2019). Although they are among the most important and common MGEs implicated in the dissemination and distribution of resistance genes between isolates (Fang et al., 2019), fewer than 10% of all isolates in the present study carried class 1 integrons, and correlation analysis showed that neither MDR nor MHMR were significantly correlated with the carrying of V. parahaemolyticus class 1 integrons.
Previous research indicated that the dissemination of antibiotic and heavy metal resistance determinants may be a result of HGT, resulting in the increased prevalence of MGEs in marine aquaculture environments (Rodriguez-Blanco et al., 2012). Among the various MGEs, class 1 integrons are thought to be strictly associated with coselection mechanisms, and harboring the class 1 integron gene can be highly beneficial for bacterial fitness (Gillings et al., 2015; Di Cesare et al., 2016). However, inconsistent with the high prevalence of MDR and MHMR, only a few class 1 integrons were detected among the isolates tested in this study. Similar results have been reported showing that antimicrobial resistance is not related to class 1, 2, or 3 integrons in Vibrio sp. isolated from seawater samples in Lima, Peru (Sulca et al., 2018). In addition, other studies have also shown a low prevalence of other MGEs, such as plasmids and integrative and conjugative elements (ICEs), in V. parahaemolyticus isolates from seafood in certain provinces in China (Hu and Chen, 2016; He et al., 2019). Thus, V. parahaemolyticus in aquatic species may have adopted other molecular mechanisms that mediate the high prevalence of resistance determinants, and this requires further study (Hu and Chen, 2016).
Conclusion
In conclusion, the results of the present study indicate that V. parahaemolyticus occurs in P. vannamei from both farms and markets regardless of whether it is freshwater-cultured or seawater-cultured. Few V. parahaemolyticus isolates were found to carry the tdh (4.44%) or trh (3.33%) toxicity genes. However, MDR isolates (40.00%) and MHMR isolates (34.44%) were commonly observed. MDR and MHMR were found to be positively correlated using Pearson’s correlation analysis (P = 0.004; R = 0.759). Most of the V. parahaemolyticus isolates represented new STs, which indicates high diversity among the isolates. However, correlation analysis revealed that neither MDR (P = 0.550; R = 0.192) nor MHMR (P = 0.990; R = 0.004) were significantly correlated with class 1 integrons in V. parahaemolyticus. Combined with the results of other studies, our findings suggest that V. parahaemolyticus in aquatic species may have adopted other molecular mechanisms that lead to the high prevalence of resistance determinants, and future research should focus on elucidating these mechanisms. The results of this study provide data to support aquatic animal health management and food safety risk assessments in the aquaculture industry.
Data Availability Statement
The datasets generated for this study can be found in the https://pubmlst.org/vparahaemolyticus, 12 novel STs (2217-2228).
Author Contributions
HJ completed MLST analysis and all data analysis and prepared the manuscript. TY, YY, and SY completed virulence gene detection, antibiotic and heavy metal resistance testing, and integron class and gene cassette detection. JW, RL, and YL completed P. vannamei sampling and V. parahaemolyticus isolation and identification. JF and CZ designed the project and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was supported by the National Natural Science Foundation of China under Grant Nos. 31901792 and 31801655 and the Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ18C200004.
References
- Ananda Raja R., Sridhar R., Balachandran C., Palanisammi A., Ramesh S., Nagarajan K. (2017). Pathogenicity profile of Vibrio parahaemolyticus in farmed pacific white shrimp, Penaeus vannamei. Fish Shellfish Immunol. 67 368–381. 10.1016/j.fsi.2017.06.020 [DOI] [PubMed] [Google Scholar]
- Ansari M. I., Grohmann E., Malik A. (2008). Conjugative plasmids in multi-resistant bacterial isolates from Indian soil. J. Appl. Microbiol. 104 1774–1781. 10.1111/j.1365-2672.2008.03736.x [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., McArthur J. V., Tuckfield R. C., Najarro M., Lindell A. H., Gooch J., et al. (2008). Antibiotic resistance in the shellfish pathogen Vibrio parahaemolyticus isolated from the coastal water and sediment of Georgia and South Carolina, USA. J. Food Prot. 71 2552–2558. 10.4315/0362-028X-71.12.2552 [DOI] [PubMed] [Google Scholar]
- Blair J. M. A., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. V. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13 42–51. 10.1038/nrmicro3380 [DOI] [PubMed] [Google Scholar]
- Bush K., Fisher J. F. (2011). Epidemiological expansion, structural studies, and clinical challenges of new beta-Lactamases from gram-negative bacteria. Annu. Rev. Microbiol. 65 455–478. 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- Chapman J. S. (2003). Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeter. Biodegr. 51, 271–276. 10.1016/S0964-8305(03)00044-1 [DOI] [Google Scholar]
- Cheng H., Jiang H., Fang J., Zhu C. (2019). Antibiotic resistance and characteristics of integrons in Escherichia coli isolated from Penaeus vannamei at a freshwater shrimp farm in Zhejiang Province, China. J. Food Prot. 82 470–478. 10.4315/0362-028X.JFP-18-444 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards for Antimicrobial Susceptibility Testing, 27th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Dahanayake P. S., Hossain S., Wickramanayake M. V. K. S., Heo G. J. (2019). Prevalence of virulence and extended-spectrum beta-lactamase (ESBL) genes harbouring Vibrio spp. isolated from cockles (Tegillarca granosa) marketed in Korea. Lett. Appl. Microbiol. (in press). 10.1111/lam.13232 [DOI] [PubMed] [Google Scholar]
- Devi R., Surendran P. K., Chakraborty K. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the southwest coast of India. World J. Microbiol. Biotechnol. 25 2005–2012. 10.1007/s11274-009-0101-8 [DOI] [Google Scholar]
- Di Cesare A., Eckert E. M., D’Urso S., Bertoni R., Gillan D. C., Wattiez R., et al. (2016). Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 94 208–214. 10.1016/j.watres.2016.02.049 [DOI] [PubMed] [Google Scholar]
- Elmahdi S., DaSilva L. V., Parveen S. (2016). Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol. 57 128–1234. 10.1016/j.fm.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Etesami H. (2018). Bacterial mediated alleviation of heavy metal stress and decreased accumulation of metals in plant tissues: mechanisms and future prospects. Ecotoxicol. Environ. Saf. 147 175–191. 10.1016/j.ecoenv.2017.08.032 [DOI] [PubMed] [Google Scholar]
- Fang J., Shen Y., Qu D., Han J. (2019). Antimicrobial resistance profiles and characteristics of integrons in Escherichia coli strains isolated from a large-scale centralized swine slaughterhouse and its downstream markets in Zhejiang, China. Food Control 95 215–222. 10.1016/j.foodcont.2018.08.003 [DOI] [Google Scholar]
- Ghosh A. K. (2018). Effect of feeding level on growth, body composition, fatty acid profile, and nutrient accumulation in shrimp (Litopenaeus vannamei). Aquac. Int. 26 405–417. 10.1007/s10499-017-0225-z [DOI] [Google Scholar]
- Gillings M. R., Gaze W. H., Pruden A., Smalla K., Tiedje J. M., Zhu Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9 1269–1279. 10.1038/ismej.2014.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Escalona N., Martinez-Urtaza J., Romero J., Espejo R. T., Jaykus L. A., DePaola A. (2008). Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190 2831–2840. 10.1128/JB.01808-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Tang H., Ren C., Wang G., Zhou L., Han C. (2015). Prevalence and genetic diversity of clinical Vibrio parahaemolyticus isolates from China, revealed by multilocus sequence typing scheme. Front. Microbiol. 6:291. 10.3389/fmicb.2015.00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Walker R. D., Janes M. E., Prinyawiwatkul W., Ge B. (2007). Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73 7096–7098. 10.1128/aem.01116-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Jin L., Sun F., Hu Q., Chen L. (2016). Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from fresh shrimps in Shanghai fish markets, China. Environ. Sci. Pollut. Res. 23 15033–15040. 10.1007/s11356-016-6614-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Wang S., Zhang J., Zhang X., Sun F., He B., et al. (2019). Integrative and conjugative elements-positive Vibrio parahaemolyticus isolated from aquaculture shrimp in Jiangsu, China. Front. Microbiol. 10:1574. 10.3389/fmicb.2019.01574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Chen L. (2016). Virulence and antibiotic and heavy metal resistance of Vibrio parahaemolyticus isolated from crustaceans and shellfish in Shanghai, China. J. Food Prot. 79 1371–1377. 10.4315/0362-028X.JFP-16-031 [DOI] [PubMed] [Google Scholar]
- Jaffer Y. D., Saraswathy R., Ishfaq M., Antony J., Bundela D. S., Sharma P. C. (2020). Effect of low salinity on the growth and survival of juvenile pacific white shrimp, Penaeus vannamei: a revival. Aquaculture 515:734561 10.1016/j.aquaculture.2019.734561 [DOI] [Google Scholar]
- Jiang Y., Chu Y., Xie G., Li F., Wang L., Huang J., et al. (2019). Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 290 116–124. 10.1016/j.ijfoodmicro.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Kadlec K., Kehrenberg C., Schwarz S. (2007). Efflux-mediated resistance to florfenicol and/or chloramphenicol in Bordetella bronchiseptica: identification of a novel chloramphenicol exporter. J. Antimicrob. Chemother. 59 191–196. 10.1093/jac/dkl498 [DOI] [PubMed] [Google Scholar]
- Kang C. H., Shin Y., Yu H., Kim S., So J. S. (2018). Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from oysters in Korea. Mar. Pollut. Bull. 135 69–74. 10.1016/j.marpolbul.2018.07.007 [DOI] [PubMed] [Google Scholar]
- Law J. W., Letchumanan V., Chan K., Goh B., Lee L. (2017). “Insights into detection and identification of foodborne pathogens,” in Foodborne Pathogens and Antibiotic Resistance, ed. Singh O. V., (Hoboken, NJ: John Wiley & Sons, Inc; ), 153–201. 10.1002/9781119139188.ch7 [DOI] [Google Scholar]
- Lee L., Ab Mutalib N., Law J. W., Wong S. H., Letchumanan V. (2018). Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in Selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front. Microbiol. 9:2513. 10.3389/fmicb.2018.02513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V., Ab Mutalib N., Wong S. H., Chan K., Lee L. (2019a). Determination of antibiotic resistance patterns of Vibrio parahaemolyticus from shrimp and shellfish in Selangor, Malaysia. Prog. Mircobes Mol. Bio1. 2:a0000019 10.36877/pmmb.a0000019 [DOI] [Google Scholar]
- Letchumanan V., Loo K., Law J. W., Wong S. H., Goh B., Ab Mutalib N., et al. (2019b). Vibrio parahaemolyticus: the Protagonist causing foodborne diseases. Prog. Mircobes Mol. Bio1. 2:a0000029 10.36877/pmmb.a0000029 [DOI] [Google Scholar]
- Letchumanan V., Chan K., Pusparajah P., Saokaew S., Duangjai A., Goh B., et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. 10.3389/fmicb.2016.01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letchumanan V., Yin W., Lee L., Chan K. (2015). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. 10.3389/fmicb.2015.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Liu J., Xu G., Chen B. (2019). Distribution and transport of heavy metals in surface sediments of the Zhejiang nearshore area, East China Sea: sedimentary environmental effects. Mar. Pollut. Bull. 146 542–551. 10.1016/j.marpolbul.2019.07.001 [DOI] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lopatek M., Wieczorek K., Osek J. (2018). Antimicrobial resistance, virulence factors, and genetic profiles of Vibrio parahaemolyticus from seafood. Appl. Environ. Microbiol. 84:e00537-18. 10.1128/AEM.00537-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Maury L., Garcia-Dominguez M., Florencio F. J., Reyes J. C. (2002). A two-component signal transduction system involved in nickel sensing in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 43 247–256. 10.1046/j.1365-2958.2002.02741.x [DOI] [PubMed] [Google Scholar]
- Luo Y., Mao D., Rysz M., Zhou Q., Zhang H., Xu L., et al. (2010). Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ. Sci. Technol. 44 7220–7225. 10.1021/es100233w [DOI] [PubMed] [Google Scholar]
- Malik A., Aleem A. (2011). Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environ. Monit. Assess. 178 293–308. 10.1007/s10661-010-1690-2 [DOI] [PubMed] [Google Scholar]
- Matyar F., Kaya A., Dincer S. (2008). Antimicrobial agents and heavy metal resistance in Gram-negative bacteria isolated from seawater, shrimp and sediment in Iskenderun bay, Turkey. Sci. Total Environ. 407 279–285. 10.1016/j.scitotenv.2008.08.014 [DOI] [PubMed] [Google Scholar]
- Mok J. S., Ryu A., Kwon J. Y., Kim B., Park K. (2019). Distribution of Vibrio species isolated from bivalves and bivalve culture environments along the Gyeongnam coast in Korea: virulence and antimicrobial resistance of Vibrio parahaemolyticus isolates. Food Control 106:106697 10.1016/j.foodcont.2019.06.023 [DOI] [Google Scholar]
- Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its Serovariants. Clin. Microbiol. Rev. 20 39–48. 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odumosu B. T., Adeniyi B. A., Chandra R. (2013). Analysis of integrons and associated gene cassettes in clinical isolates of multidrug resistant Pseudomonas aeruginosa from Southwest Nigeria. Ann. Clin. Microbiol. Antimicrob. 12:29. 10.1186/1476-0711-12-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani D., Leoni F., Talevi G., Masini L., Santarelli S., Rocchegiani E., et al. (2013). Extensive investigation of antimicrobial resistance in Vibrio parahaemolyticus from shellfish and clinical sources, Italy. Int. J. Antimicrob. Agents 42 191–193. 10.1016/j.ijantimicag.2013.05.003 [DOI] [PubMed] [Google Scholar]
- Peng M., Liu X., Niu D., Ye B., Lan T., Dong Z., et al. (2019). Survival, growth and physiology of marine bivalve (Sinonovacula constricta) in long-term low-salt culture. Sci. Rep. 9:2819. 10.1038/s41598-019-39205-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25 820–832. 10.1016/j.tim.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Raghunath P., Acharya S., Blianumathi A., Karunasagar I., Karunasagar I. (2008). Detection and molecular characterization of Vibrio parahaemolyticus isolated from seafood harvested along the southwest coast of India. Food Microbiol. 25 824–830. 10.1016/j.fm.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Blanco A., Lemos M. L., Osorio C. R. (2012). Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob. Agents Chemother. 56 2619–2626. 10.1128/AAC.05997-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C., Berendonk T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. 10.3389/fmicb.2012.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva I. P., Carneiro C. D. S., Fontes Saraiva M. A., Santos de Oliveira T. A., de Sousa O. V., Evangelista-Barreto N. S. (2018). Antimicrobial resistance and potential virulence of Vibrio parahaemolyticus isolated from water and bivalve mollusks from Bahia, Brazil. Mar. Pollut. Bull. 131 757–762. 10.1016/j.marpolbul.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Silvester R., Alexander D., Ammanamveetil M. H. A. (2015). Prevalence, antibiotic resistance, virulence and plasmid profiles of Vibrio parahaemolyticus from a tropical estuary and adjoining traditional prawn farm along the southwest coast of India. Ann. Microbiol. 65 2141–2149. 10.1007/s13213-015-1053-x [DOI] [Google Scholar]
- Sulca M. A., Orozco R., Alvarado D. E. (2018). Antimicrobial resistance not related to 1,2,3 integrons and superintegron in Vibrio spp. isolated from seawater sample of Lima (Peru). Mar. Pollut. Bull. 131 370–377. 10.1016/j.marpolbul.2018.04.050 [DOI] [PubMed] [Google Scholar]
- Tan C. W., Malcolm T. T. H., Kuan C. H., Thung T. Y., Chang W. S., Loo Y. Y., et al. (2017). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from Short Mackerels (Rastrelliger brachysoma) in Malaysia. Front. Microbiol. 8:1087. 10.3389/fmicb.2017.01087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. T., Lee L., Goh B. (2020). Critical review of fermentation and extraction of anti-Vibrio compounds from Streptomyces. Prog. Mircobes Mol. Bio1. 3:a0000051 10.36877/pmmb.a0000051 [DOI] [Google Scholar]
- Tetens J. L., Billerbeck S., Schwenker J. A., Hoelzel C. S. (2019). Short communication: selection of extended-spectrum beta-lactamase-producing Escherichia coli in dairy calves associated with antibiotic dry cow therapy-A cohort study. J. Dairy Sci. 102 11449–11452. 10.3168/jds.2019-16659 [DOI] [PubMed] [Google Scholar]
- Theethakaew C., Feil E. J., Castillo-Ramirez S., Aanensen D. M., Suthienkul O., Neil D. M., et al. (2013). Genetic relationships of Vibrio parahaemolyticus isolates from clinical, human carrier, and environmental sources in Thailand, determined by multilocus sequence analysis. Appl. Environ. Microbiol. 79 2358–2370. 10.1128/AEM.03067-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urmersbach S., Alter T., Koralage M. S., Sperling L., Gerdts G., Messelhausser U., et al. (2014). Population analysis of Vibrio parahaemolyticus originating from different geographical regions demonstrates a high genetic diversity. BMC Microbiol. 14:59. 10.1186/1471-2180-14-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. H. Y., Liu M., Wan H. Y., Chen S. (2012). Characterization of extended-spectrum-beta-lactamase-producing Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 56 4026–4028. 10.1128/AAC.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T., Wu Q., Zhang J., Xu X., Cheng J. (2017). Comparison of Vibrio parahaemolyticus isolates from aquatic products and clinical by antibiotic susceptibility, virulence, and molecular characterisation. Food Control 71 315–321. 10.1016/j.foodcont.2016.06.046 [DOI] [Google Scholar]
- Xie T., Xu X., Wu Q., Zhang J., Cheng J. (2016). Prevalence, molecular characterization, and antibiotic susceptibility of Vibrio parahaemolyticus from Ready-to-Eat foods in China. Front. Microbiol. 7:549. 10.3389/fmicb.2016.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Li E., Liu Y., Wang S., Wang X., Chen K., et al. (2018). Effect of dietary lipid level on growth, lipid metabolism and health status of the Pacific white shrimp Litopenaeus vannamei at two salinities. Aquac. Nutr. 24 204–214. 10.1111/anu.12548 [DOI] [Google Scholar]
- Xu X., Cheng J., Wu Q., Zhang J., Xie T. (2016). Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol. 16:32. 10.1186/s12866-016-0650-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Guan W., Xie D., Lu W., Ren X., Yuan J., et al. (2019). Evaluation of immunological response in shrimp Penaeus vannamei submitted to low temperature and air exposure. Dev. Comp. Immunol. 100:103413. 10.1016/j.dci.2019.103413 [DOI] [PubMed] [Google Scholar]
- Yang F., Yang F., Wang G., Kong T., Wang H., Zhang C. (2020). Effects of water temperature on tissue depletion of florfenicol and its metabolite florfenicol amine in crucian carp (Carassius auratus gibelio) following multiple oral doses. Aquaculture 515:734542 10.1016/j.aquaculture.2019.734542 [DOI] [Google Scholar]
- Yang Y., Xie J., Li H., Tan S., Chen Y., Yu H. (2017). Prevalence, antibiotic susceptibility and diversity of Vibrio parahaemolyticus isolates in seafood from South China. Front. Microbiol. 8:2566. 10.3389/fmicb.2017.02566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano Y., Hamano K., Satomi M., Tsutsui I., Aue-umneoy D. (2011). Diversity and characterization of oxytetracycline-resistant bacteria associated with non-native species, white-leg shrimp (Litopenaeus vannamei), and native species, black tiger shrimp (Penaeus monodon), intensively cultured in Thailand. J. Appl. Microbiol. 110 713–722. 10.1111/j.1365-2672.2010.04926.x [DOI] [PubMed] [Google Scholar]
- Yassin A. K., Gong J., Kelly P., Lu G., Guardabassi L., Wei L., et al. (2017). Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock, China. PLoS One 12:e0185326. 10.1371/journal.pone.0185326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Niu M., Yu M., Liu Y., Wang D., Shi X. (2016). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shellfish in Shanghai. Food Control 60 263–268. 10.1016/j.foodcont.2015.08.005 [DOI] [Google Scholar]
- Zhao S., Wei W., Fu G., Zhou J., Wang Y., Li X., et al. (2020). Application of biofertilizers increases fluoroquinolone resistance in Vibrio parahaemolyticus isolated from aquaculture environments. Mar. Pollut. Bull. 150:110592. 10.1016/j.marpolbul.2019.110592 [DOI] [PubMed] [Google Scholar]
- Zimmerman A. M., DePaola A., Bowers J. C., Krantz J. A., Nordstrom J. L., Johnson C. N., et al. (2007). Variability of total and pathogenic Vibrio parahaemolyticus densities in northern Gulf of Mexico water and oysters. Appl. Environ. Microbiol. 73 7589–7596. 10.1128/aem.01700-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the https://pubmlst.org/vparahaemolyticus, 12 novel STs (2217-2228).