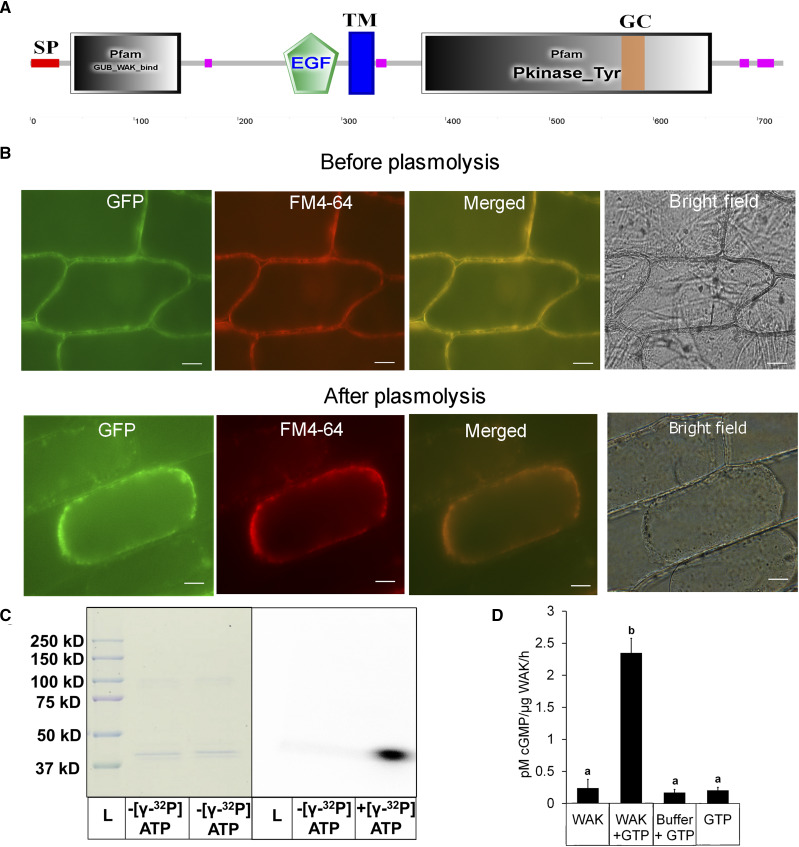
Figure 5.
Biochemical characterization and localization of OsWAKL21.2. A, Domain architecture of OsWAKL21.2 using the SMART tool (http://smart.embl-heidelberg.de/). EGF, Epidermal growth factor-like repeat; GC, GC motif; GUB, galacturonan-binding domain; Pkinase_Tyr, kinase domain; SP, signal peptide; TM, transmembrane region. Pink boxes represent low-complexity regions as indicated by the SMART tool. B, OsWAKL21.2:EGFP localizes on the cell membrane in onion peel after transient expression. OsWAKL21.2:EGFP was transiently transformed to onion peel cells using A. tumefaciens, and peels were visualized after 2 d using an epifluorescence microscope. Plasmolysis was done by incubating onion peel in 0.8 m mannitol for 30 min. The experiment was repeated three times, and similar results were obtained. FM4-64 staining was done by incubating in 10 μg mL−1 dye for 10 min. Images were captured at 60×. Bars = 10 μm. C, Kinase assay. The kinase domain of OsWAKL21 cloned and purified from E. coli shows autophosphorylation activity. Fifty micrograms of affinity-purified recombinant protein was used for assays with or without radiolabeled ATP. After 1 h, denatured samples were loaded onto a 10% SDS-PAGE gel. The gel was further subjected to autoradiography and Coomassie Brilliant Blue staining. The experiment was repeated three times, and similar results were obtained. D, GC assay. Fifty micrograms (in 50 μL) of affinity-purified recombinant protein was used for GC assay with or without GTP. After 1 h, 5 μL of the sample was directly used for cGMP quantification. GTP alone and GC buffer + GTP were used as controls. Each bar indicates the average, and error bars represent se of three independent experiments. Lowercase letters (a and b) above the bars indicate significant differences when analyzed using Student’s t test with P < 0.05.