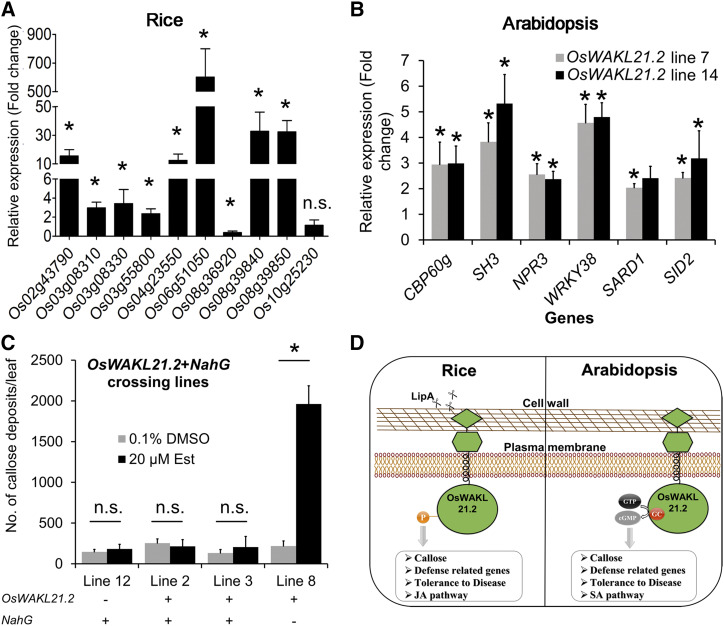
Figure 8.
OsWAKL21.2 induces the expression of JA pathway-related genes in rice, whereas it activates SA pathway-related genes in Arabidopsis. A, Relative expression of 10 JA pathway-related genes after transient overexpression of OsWAKL21.2 in rice leaves. These genes include three ZIM domain-containing proteins (LOC_Os03g08310, LOC_Os03g08330, and LOC_Os10g25230), two lipoxygenases (LOC_Os08g39840 and LOC_Os08g39850), one allene oxide synthase (LOC_Os03g55800), one basic helix-loop-helix transcription factor (LOC_Os04g23550), one ethylene-responsive transcription factor (LOC_Os02g43790), one chitinase (LOC_Os06g51050), and an AP2 domain-containing transcription factor (LOC_Os08g36920). For each gene, the transcript level under uninduced conditions (treatment with A. tumefaciens carrying WAK-wt [wild type] with 0.1% [v/v] DMSO) was considered as 1 and was compared with that under the induced conditions (treatment with A. tumefaciens carrying WAK-wt with 20 µm Est). Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 12 in each experiment). OsACTIN1 was used as an internal control. The relative FC was calculated by using the 2−∆∆Ct method. Asterisks represent significant differences in FC using Student’s t test with P < 0.05. n.s. indicates no significant difference in relative expression. B, Effects of heterologous expression of OsWAKL21.2 on the expression of SA pathway-related genes in transgenic Arabidopsis lines. Expression in 0.1% (v/v) DMSO-treated leaves was considered as 1, and relative expression in 20 µm Est-treated leaves was calculated with respect to it. Each bar represents the average FC, and error bars indicate se of three independent experiments (n = 3 in each experiment). AtACTIN2 was used as an internal control for RT-qPCR. The relative FC was calculated by using the 2−∆∆Ct method. Asterisks represent significant differences in FC using Student’s t test with P < 0.05. C, Quantification of callose deposits in Arabidopsis crossing lines expressing NahG and OsWAKL21.2 (lines 2 and 3) or either one of those (NahG only, line 12; OsWAKL21.2-wt only, line 8). Leaves were treated with either 20 µm Est (inducer) or 0.1% (v/v) DMSO (control). Each bar represents the average, and error bars represent se of three leaves in an experiment. The asterisk represents a significant difference when calculated using one-way ANOVA followed by the Tukey-Kramer multiple comparison test with P < 0.05. n.s. indicates no significant difference. D, Model depicting the predicted role of OsWAKL21.2 in the induction of immune responses in rice and Arabidopsis. OsWAKL21.2 seems to be either directly or indirectly involved in the activation of immune responses following treatment with LipA. The kinase activity of OsWAKL21.2 is required for the induction of immune responses and the activation of the JA pathway in rice. On the other hand, in Arabidopsis, the GC activity of OsWAKL21.2 is required for the induction of immune responses that appear to be through the SA pathway.