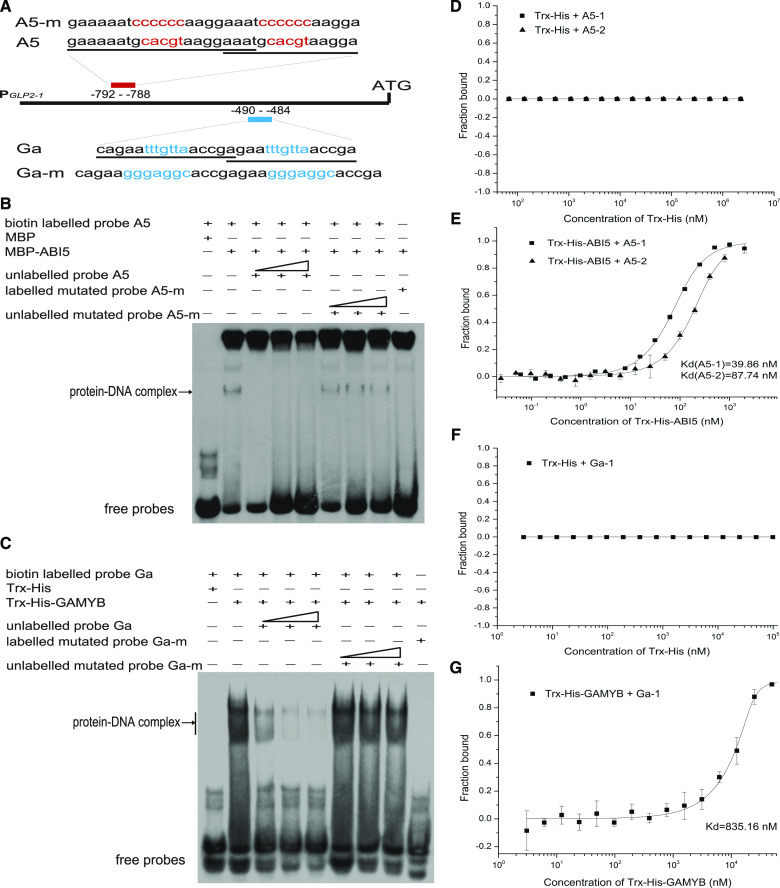
Figure 6.
Physical interaction of ABI5 and GAMYB to the ABRE and GARE in PGLP2-1. A to C, EMSA results displayed that the transcription factors (ABI5 and GAMYB) could directly recognize and bind to PGLP2-1 fragments containing the ABRE or GARE, respectively. A, The probes used in EMSA were designed based on the predicted binding sequence of ABRE (red letters) and GARE (blue letters) in the F1 and F2 fragments, respectively, together with their flanking nucleotides (underlined), two overlapped binding sites were synthesized. B and C, Binding activity of ABI5 and GAMYB with their corresponding probes. The fusion proteins MBP-ABI5 and Trx-His-GAMYB specifically bound to the corresponding probes and resulted in the retardant band, whereas no binding activity was observed with the corresponding negative controls of MBP and Trx-His. The unlabeled cold probes (10, 50, 100×) gradually competed for the corresponding binding activity, whereas the mutated biotin-labeled probes (10, 50, 100×) completely abolished the protein-DNA interaction, and the mutated cold-probes showed no competitive activities. D to G, MST analysis showing that the specially binding activity of Trx-His-ABI5 with OsGLP2-1 promoter fragments of A5-1, A5-2 (E), and Trx-His-GAMYB with Ga-1 (G), respectively. The negative control, Trx-His, did not interact with these fragments (D and F). D and E, MST measurements of the interaction between ABI5 and A5-1, A5-2 fragments. F and G, MST assay testing the interaction between GAMYB and Ga-1 fragment. D to G data are average ± sd, n = 3. The data were analyzed with the software of MO. Affinity Analysis v2.2.4.