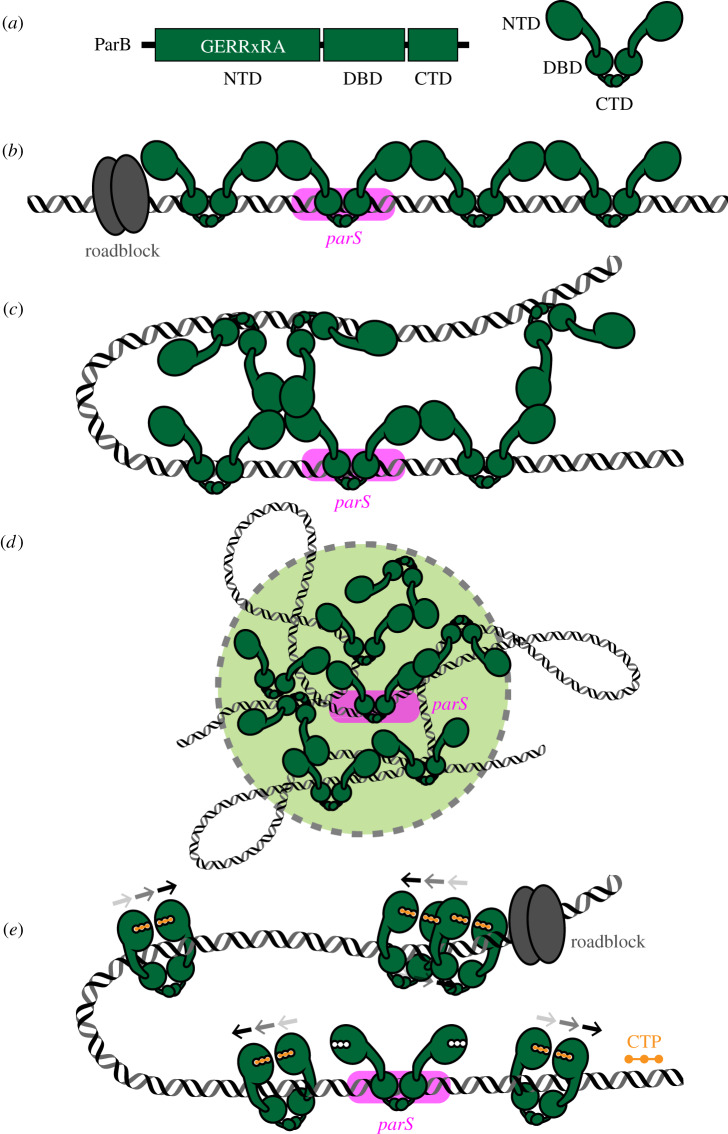
Figure 1.
The assembly of a higher-order ParB–DNA nucleoprotein complex. (a) Chromosomal ParB proteins share a common domain architecture, consisting of an N-terminal domain (NTD), a central DNA-binding domain (DBD) and a C-terminal domain (CTD). The NTD harbours a conserved arginine-rich motif (GERRxRA) that mediates ParB–ParB and ParB–cytidine triphosphate (CTP) interactions. (b) Model 1: ParB spreading by a one-dimensional filamentation. (c) Model 2: ParB spreading by bridging and condensing DNA. (d) Model 3: ParB spreading by caging DNA. (e) Model 4: ParB spreading by sliding on DNA. ParB switches from an open to a closed clamp upon binding to CTP (orange). ParB and parS are coloured green and magenta, respectively. The arrows above the ParB–CTP complexes (e) indicate their progressive sliding on DNA. A tight DNA-binding protein (grey) can unidirectionally block the one-dimensional filamentation or the sliding of ParB on DNA.