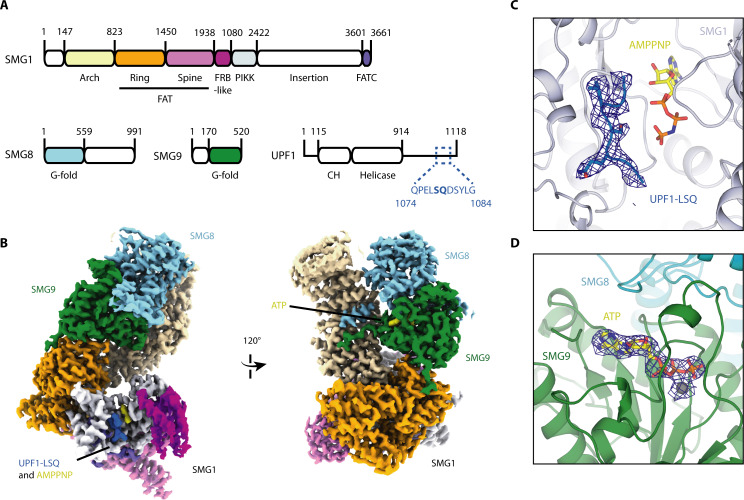
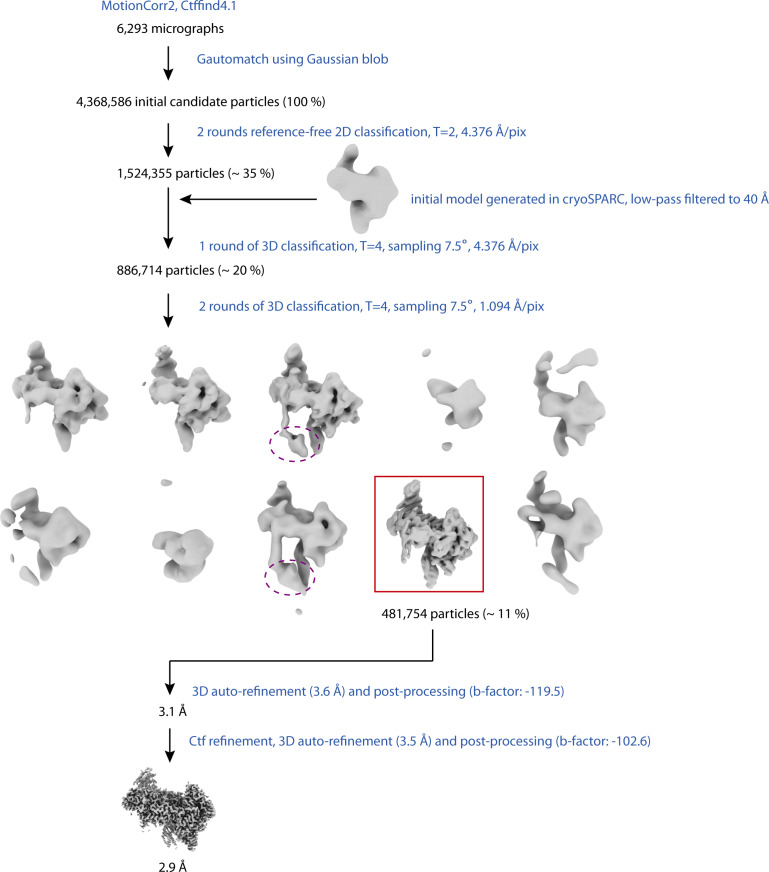
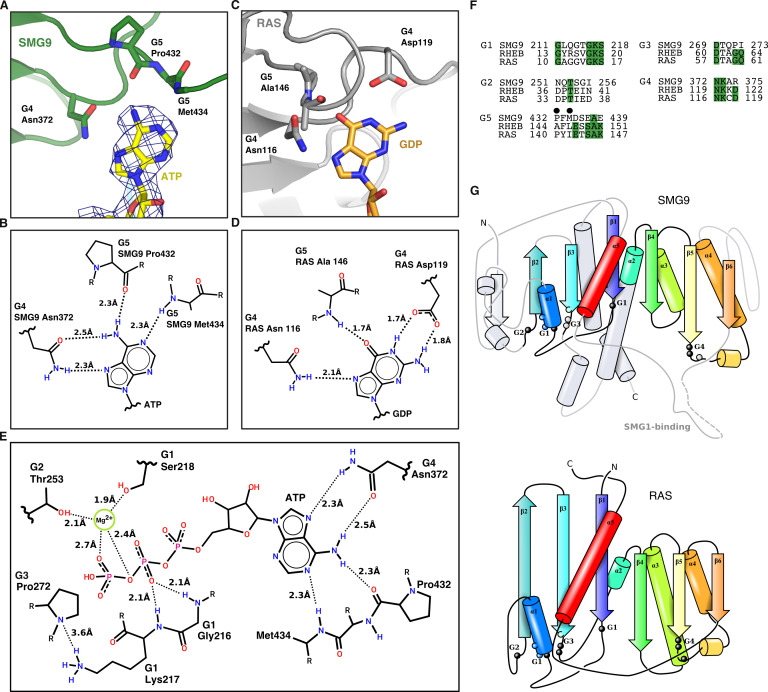
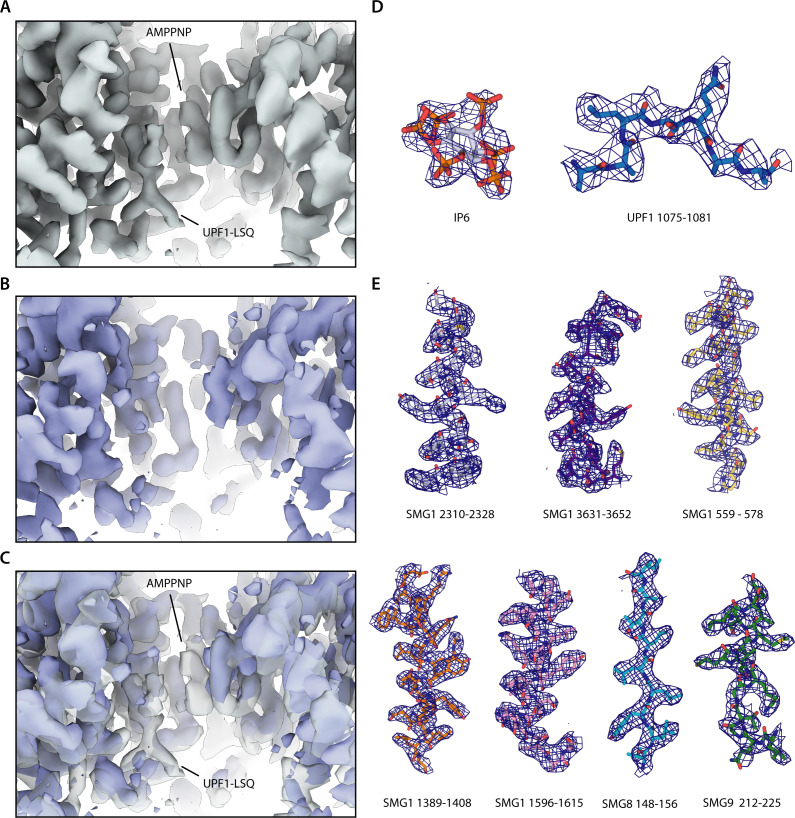
(
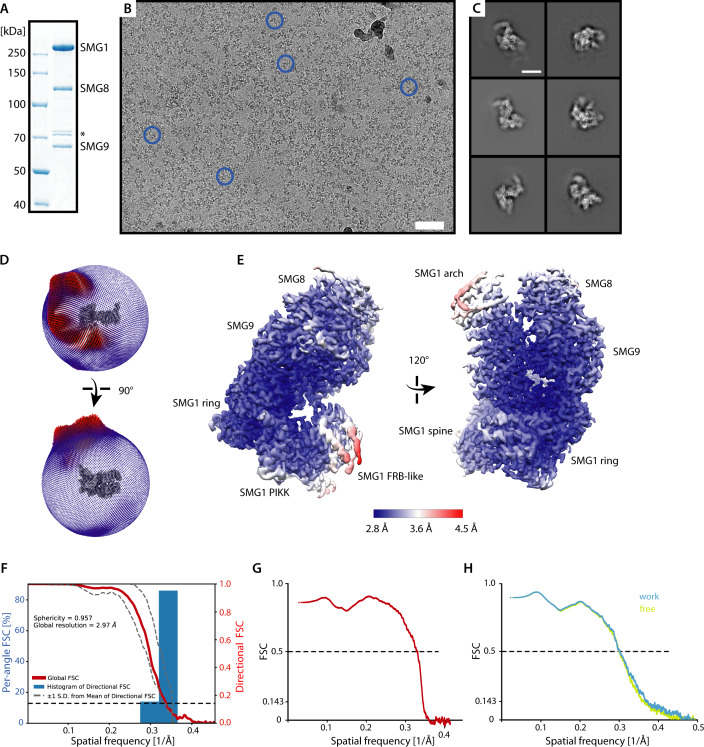
A) Coomassie-stained SDS-PAGE showing purified SMG1-8-9. The asterisk indicates contaminants. (
B) Representative micrograph of the collected data set with some SMG1-8-9 particles indicated by blue circles. Scale bar ≈ 500 Å. (
C) Representative 2D averages of picked particles. Scale bar ≈ 100 Å. (
D) Spherical angular distribution of particles contributing to the final reconstruction with larger red rods indicating more prominent particle views and smaller blue rods indicating rarer particle views. (
E) Map of SMG1-8-9 colored according to estimated local resolution shown in two different views as in
Figure 1B. Large parts of the complex including the kinase domain and the bound UPF1 peptide are resolved to around 3 Å. Important features of the map are indicated. (
F) Three-dimensional FSC plot and further analysis of orientation bias. The red line represents the estimated global masked half-map FSC curve indicating a nominal overall resolution of 2.97 Å according to the gold standard FSC cut off of 0.143 (
Rosenthal and Henderson, 2003). The spread of directional resolution values is defined as ± 1σ (shown as dashed grey lines). Overall isotropy of the map is confirmed by a sphericity of 0.957 (out of 1) (
Tan et al., 2017). (
G) Model vs. map FSC plot for the real spaced refined model. (H) Model vs. map FSC plots for half map 1 ("work") used for real space refinement after displacing atoms of the final model (σ = 0.5 Å) and half map 2 not used for refinement ("free"). The good agreement of the two curves indicates that no substantial overfitting took place.