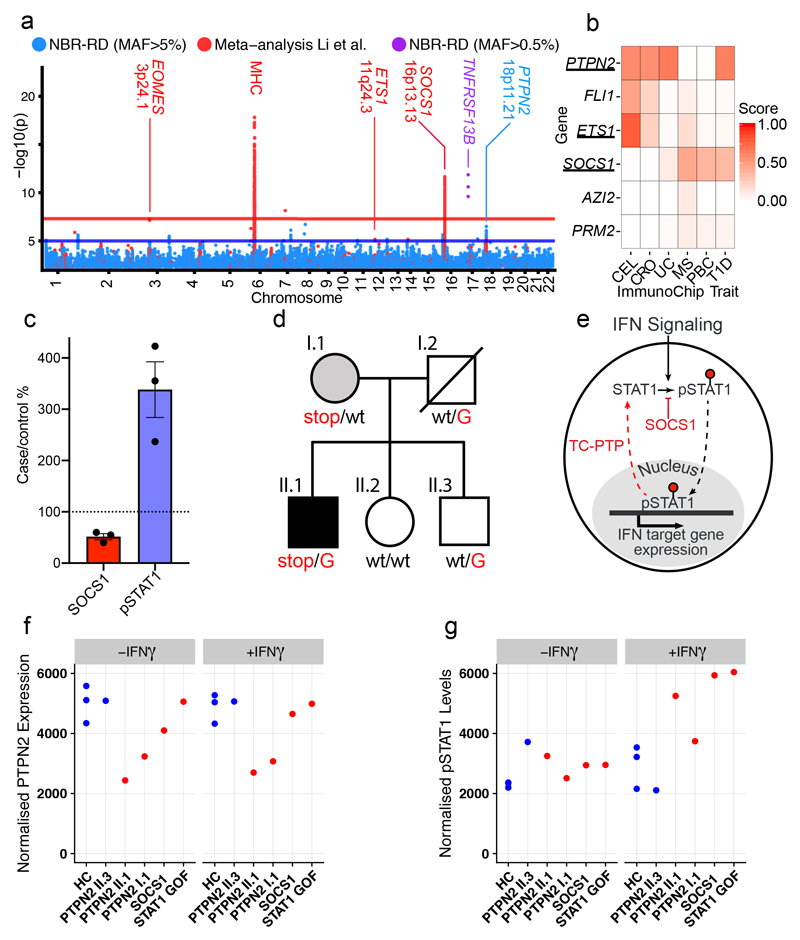
Figure 4. Antibody deficiency (AD-PID) GWAS identifies common variants that mediate disease risk and suggests novel monogenic candidate genes.
(a) A composite Manhattan plot for the AD-PID GWAS. Blue – common variants (MAF>0.05) analysed in this study (NBR-RD) only (cases n=773, controls n=9,225), red – variants from fixed effects meta-analysis with data from Li et al. (cases n=1,511, controls n=20,224); and purple – genome-wide significant low frequency (0.005<MAF<0.05) variants in TNFRSF13B locus. Loci of interest are labelled with putative causal protein coding gene names. (b) COGS prioritisation scores of candidate monogenic causes of PID using previous autoimmune targeted genotyping studies (Supplementary Table 4) across suggestive AD-PID loci (n=4). For clarity, only diseases prioritising one or more genes are shown. CEL – coeliac disease, CRO- Crohn’s disease, UC – ulcerative colitis, MS – multiple sclerosis, PBC – primary biliary cirrhosis and T1D – type 1 diabetes (c) Graph of relative pSTAT1 and SOCS1 in lysates made from 2 hour IFN-γ treated T cell blasts from SOCS1 mutation patients and controls. (Lines present mean, error bars=S.E.M.) (d) The pedigree of the PTPN2 mutation patient. Carriers of the rs2847297-G risk allele are indicated. (e) Simplified model of how SOCS1 and TC-PTP limit the phosphorylated-STAT1 triggered by interferon signalling. (f) Graph of relative PTPN2 and pSTAT1 from the indicated patients and controls, in lysates made from T cell blasts incubated ± IFN-γ for 2 hours. (PTPN2 normalized to tubulin level, pSTAT1 normalised to STAT1 levels, representative of 2 independent experiments)