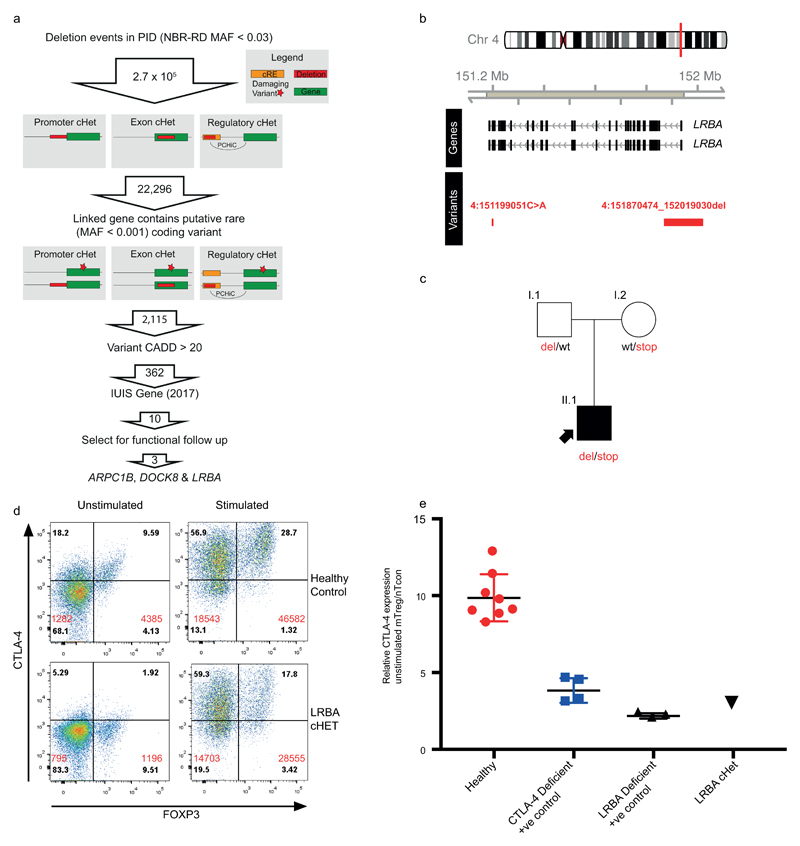
Extended Data Figure 4. Candidate cHET filtering strategy and LRBA patient.
(a) Filtering strategy to identify candidate compound heterozygous (cHET) pathogenic variants consisting of a rare coding variant in a PID-associated gene and a deletion of a cis-regulatory element for the same gene. (b) Regional plot of the compound heterozygous variants. Gene annotations for are taken from Ensembl Version 75, and the transcripts shown are those with mRNA identifiers in RefSeq (ENST00000357115 and ENST00000510413). The position of each variant relative to the gene transcript is shown by a red bar, with the longer bar indicating the extent of the deleted region. Variant coordinates are shown for the GRCh37 genome build. (c) Pedigree of LRBA patient demonstrating phase of the causal variants. (d) FACS dotplot of CTLA-4 and FoxP3 expression in LRBA cHET patient and a healthy control (representative of 2 independent experiments). Numbers in black are the percentage in each quadrant. Numbers in red are the MFI of CTLA-4 staining in FoxP3 -ve and FoxP3 +ve cells. (e) Normalised CTLA-4 expression, assessed as previously described in Hou et al. (Blood, 2017), in the LRBA cHET patient (n=1), healthy controls (n=8) and positive control CTLA-4 (n=4) and LRBA (n=3 deficient patients. Horizontal bars indicate mean +/- SEM.