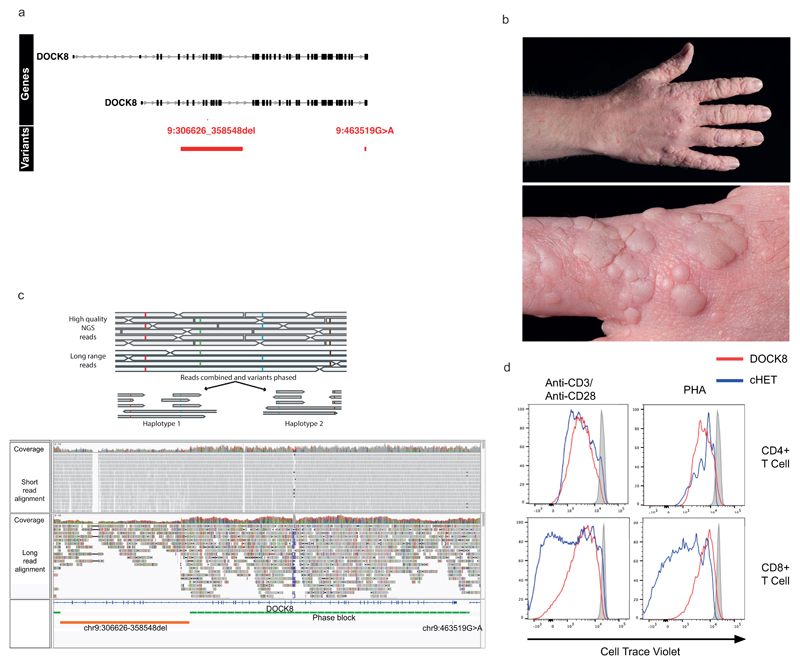
Extended Data Figure 5. DOCK8 cHET patient.
(a) Regional plot of the compound heterozygous variants. Gene annotations for are taken from Ensembl Version 75, and the transcripts shown are those with mRNA identifiers in RefSeq (ENST00000432829 and ENST00000469391). The position of each variant relative to the gene transcript is shown by a red bar, with the longer bar indicating the extent of the deleted region. Variant coordinates are shown for the GRCh37 genome build. (b) Photographs of the extensive HPV associated wart infection in the DOCK8 cHET patient. (c) cHET variant phasing. Top: cartoon representation of phasing using high quality heterozygous calls from short read WGS data and long-read nanopore sequencing data. Bottom panel: WGS and nanopore data from the DOCK8 patient. The two variants (large deletion and missense substitution) are shown in the bottom track (orange), and a single phase block (green) that spans the entire region between the two variants confirmed them to be in-trans. (d) Dye-dilution proliferation assessment in response to phytohaemagglutinin (PHA) and anti-CD3/28 beads in CD4+ and CD8+ T cells in patient and control cells (representative of 2 independent experiments). Staining was performed with CFSE dye (Invitrogen, Carlsbad, CA, USA) with the same additional fluorochrome markers as described in the flow cytometry methods section.