Figure 2: Functional cooperation between constitutive trisomy 21 and KRASG12D.
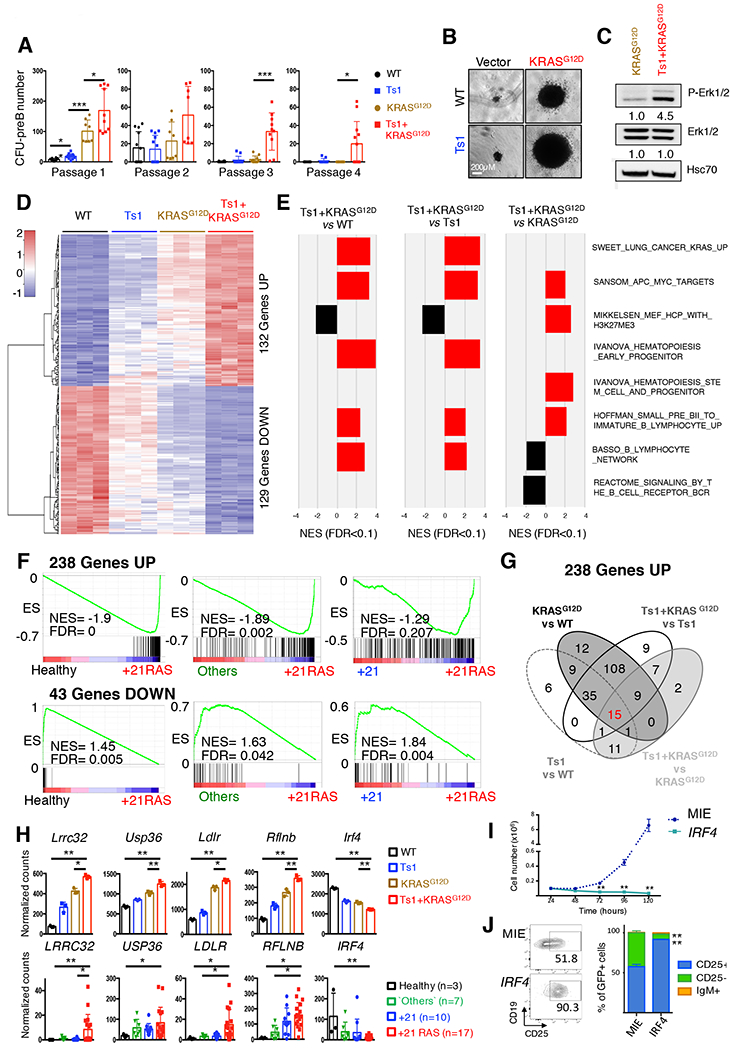
A. Number of CFU-preB colonies obtained per genotype (wild-type: WT and Ts1Rhr: Ts1) transduced with retroviral particles encoding KRASG12D or empty vector across four passages (n=8–10 replicates from 3 independent experiments; *p<0.05, **p<0.01 and ***p<0.001). B. Representative picture of the CFU-preB colonies observed at passage 1 in WT, Ts1, KRASG12D and Ts1-KRASG12D (scale bars, 0.2 mm). C. Western blot assessing the constitutive phosphorylation of Erk1/2 in starved WT-KRASG12D and Ts1Rhr-KRASG12D cells. Quantifications of the band relative intensities are indicated. D. Heatmap representing the expression of the 261 genes commonly deregulated in the WT, Ts1, KRASG12D and Ts1+KRASG12D murine B cell progenitors CFU-preB (132 Upregulated and 129 downregulated genes), n= 3 replicates per condition. E. Gene set enrichment analyses (GSEA) of selected enriched datasets; all FDR < 0.1, see also Supplemental Figure 4 and Table 6. F. GSEA of the 238 upregulated and 43 downregulated genes across subgroups of human B-ALL contained in our cohort: Others, +21 (gain of chromosome 21, no RAS alterations) and +21RAS (gain of chromosome 21 with N/KRAS mutations). G. Venn diagram assessing the association of the 238 upregulated genes among the murine paired comparisons Ts1 vs WT, KRASG12D vs WT, Ts1+KRASG12D vs Ts1 and Ts1+KRASG12D vs KRASG12D. The same analysis was performed with the 43 downregulated genes, leading to the identification of 18 ‘cooperative’ genes (see Supplemental Table 7). H. Normalized expression of five selected ‘cooperative’ genes, commonly deregulated in murine CFU-preB colonies (WT, Ts1, KRASG12D, Ts1+KRASG12D, upper panel) and in human primary B-ALL samples (lower panel). *p<0.05 and **p<0.001. I. Growth of sorted GFP-positive Ts1+KRASG12D ectopically expressing IRF4 compared to empty vector (MIE). **p<0.001. J. Representative dot plots (left panel) and associated bar graphs (right panel, n=3) showing acquisition of CD25 surface marker in IRF4-overexpressing Ts1+KRASG12D cells (at 48h). **p<0.001.
