Figure 6. HDAC5 is upregulated after inhibition of KRAS*.
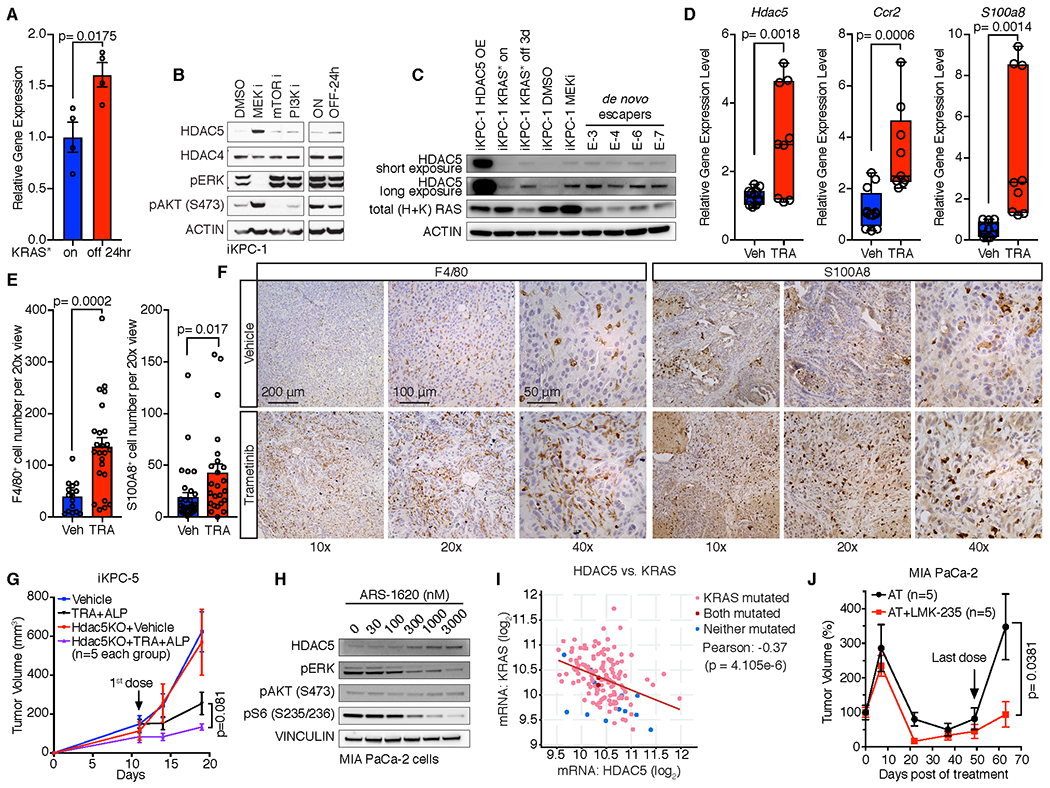
A, Hdac5 expression in KRAS*-expressing iKPC tumors and tumors after KRAS* extinction for 24 hours. B, Western blot analysis of HDAC5 expression in iKPC-1 cells following treatment with DMSO control, MEK inhibitor (Trametinib, 50nM), PI3K inhibitor (Ly294002, 2 μM) and mTOR inhibitor (Rapamicin, 100nM), and in iKPC-1 cells w/ and w/o DOX treatment for 24 hours. C, Western blot analysis of HDAC5 protein levels in HDAC5-OE iKPC-1 cells, KRAS* on and off iKPC-1 cells, MEK inhibited iKPC-1 cells, and four de novo generated escaper cells. D-F, Comparison of mRNA expression of Hdac5, S100a8 and Ccr2 (D), quantification of F4/80+ and S100A8+ cells (E), and IHC analysis of F4/80 and S100A8 (F) in orthotopically transplanted iKPC-5 tumors treated with vehicle control (n = 4) or Trametinib (n = 3, 0.3 mg/kg, oral, daily) in C57BL/6 mice. For E, eight images were taken for each tumor and counted, and data are represented as mean ± SEM. G, Knockout of Hdac5 in combination with MEK inhibitor Trametinib (TRA) and PI3Kα inhibitor Alpelisib (ALP) impaired subcutaneously transplanted iKPC-5 tumor growth in nude mouse (n = 5). H, Western blot analysis of HDAC5 expression after treatment with KRASG12C inhibitor ARS-1620 in human MIA PaCa-2 PDAC cells. I, Correlation analysis between HDAC5 and KRAS mRNA expression in TCGA PAAD dataset by cBioPortal. The p value was calculated by two-sided t-test. J, Comparison of MIA PaCa-2 subcutaneous xenograft tumor growth between treatment with dual inhibitor combination of ARS-1620 (200 mg/kg, oral, q.d.) and Trametinib (1 mg/kg, oral, q.d.) and triple inhibitor combination of ARS-1620 (200 mg/kg, oral, q.d.), Trametinib (1 mg/kg, oral, q.d.) and LMK-235 (5 mg/kg, i.p., q.d.) in nude mice. For A and E, data are represented as mean ± SEM. For A, D, E, G and J, two-tailed unpaired t tests were performed to calculate the p values.
