Extended Data Fig. 7. gnomAD-SV can augment disease association studies.
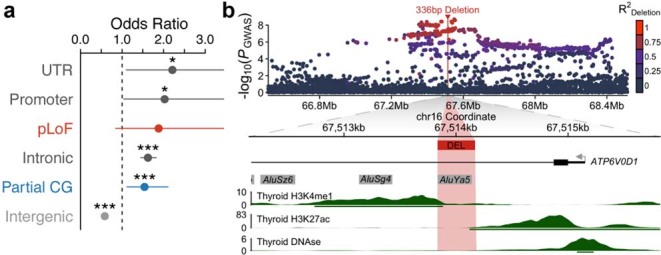
a, Functional enrichments of 2,307 common SVs in strong linkage disequilibrium (R2 ≥ 0.8) with an SNV associated with a trait or disease in the GWAS catalogue or the UK Biobank33,34. Points represent odds ratios of SVs being in strong linkage disequilibrium with at least one GWAS-significant SNV among all SVs in strong linkage disequilibrium with at least one SNV (total n = 15,634 SVs). Single and triple asterisks correspond to nominal (P < 0.05) and Bonferroni-corrected (P < 0.0083) significance thresholds from a two-sided Fisher’s exact test, respectively. Bars represent 95% confidence intervals. Test statistics, SV counts, and P values are provided in Supplementary Table 6. b, Example locus at 16q22.1, where we identified a 336-bp deletion in strong linkage disequilibrium with SNVs significantly associated with hypothyroidism in the UK Biobank34. Top, the GWAS signal among genotyped SNVs in the UK Biobank, coloured by strength of linkage disequilibrium (Pearson’s R2 value) with the 336-bp deletion identified in gnomAD-SV. Bottom, the local genomic context of this deletion, which overlaps an annotated intronic Alu element near (<1 kb) the first exon of a highly constrained, thyroid-expressed gene, ATP6V0D1. The deletion lies amidst histone mark peaks commonly found at active enhancers (H3K27ac and H3K4me1) based on publicly available chromatin data from adult thyroid samples, a phenotype-relevant tissue48. Human Alu elements are known to frequently act as enhancers, and the sentinel hypothyroidism SNV from the UK Biobank GWAS is a significant expression-modifying variant (that is, eQTL) for ATP6V0D1 and other nearby genes across many tissues, which indicates that the hypothyroidism risk haplotype modifies expression of ATP6V0D1 and/or other genes, potentially through the deletion of an intronic enhancer4,49.
