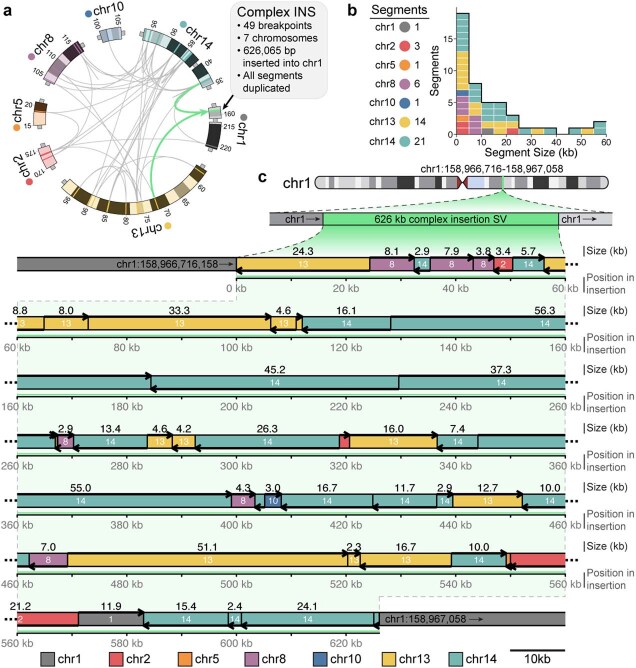
Extended Data Fig. 8. An extremely complex SV involving 49 breakpoints and seven chromosomes.
A highly complex insertion rearrangement from gnomAD-SV in which 47 segments from six different chromosomes were duplicated and inserted into a single locus on chromosome 1, forming a 626,065 bp stretch of contiguous inserted sequence composed of shattered fragments. Given the involvement of multiple chromosomes, the signature of localized shattering, and the clustered breakpoints, we note that this rearrangement has several hallmarks of germline chromothripsis, which has been observed in healthy adults previously, albeit rarely22. However, unlike previous reports of germline chromothripsis, there are no apparent whole-chromosome translocations, and all segments were duplicated before being inserted in a compound manner into chromosome 1, potentially suggesting a replication-based repair mechanism. The exact origin of this rearrangement is unclear. a, Circos representation of all 49 breakpoints and seven chromosomes involved in this SV. Teal arrows indicate insertion point into chromosome 1. b, The median segment size was 8.4 kb. c, Linear representation of the rearranged inserted sequence. Colours correspond to chromosome of origin, and arrows indicate strandedness of the inserted sequence, relative to the GRCh37 reference.