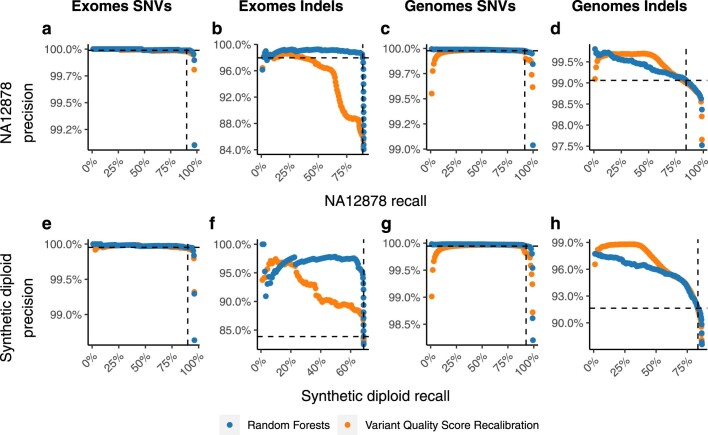
Extended Data Fig. 2. Variant calling performance for common variants.
a–h, Precision-recall curves are shown for variant calls in two samples with independent gold-standard data, NA1287849 (a–d) and a synthetic diploid mixture50 (e–h). The random forest (blue) approach described here is compared to the current state-of-the-art GATK variant quality score recalibration (orange) for exome SNVs (a, e) and indels (b, f), and genome SNVs (c, g) and indels (d, h). Note that the indels presented in f and h exclude 1-base-pair (bp) indels as they are not well characterized in the synthetic diploid mixture gold standard sample. In all cases, at the thresholds chosen (dashed lines representing 10% and 20% of SNVs and indels filtered, respectively), random forest outperforms or is similar to variant quality score recalibration.