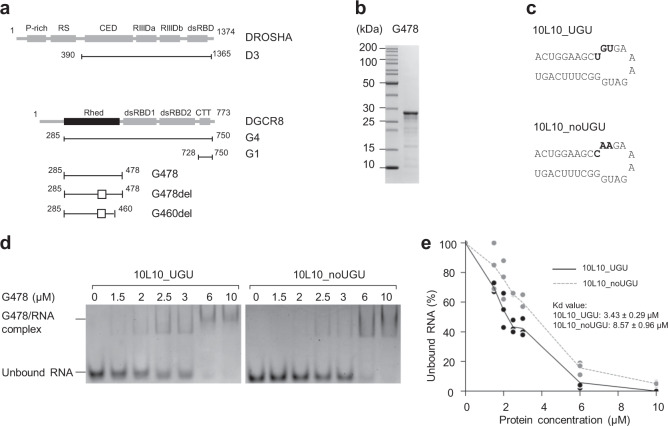
Fig. 1. Rhed recognizes the UGU motif.
a The protein constructs used in this study. For each construct, the first and last amino acid residue positions are shown. The box marks the deleted regions from amino acids 370–429. P-rich: proline-rich domain, RS: arginine/serine-rich domain, CED: central domain, RIIIDa and RIIIDb: RNase III domains, dsRBD: dsRNA-binding domain, Rhed: RNA-binding heme domain, and CTT: C-terminal tail region. b SDS-PAGE to show the purified G478 protein. c Structure diagrams and ribonucleotide sequences of 10L10_UGU and 10L10_noUGU. d The EMSAs for G478. Various amounts of G478 (ranging from 0 to 10 μM) were mixed with 1 μM of either 10L10_UGU or 10L10_noUGU in a 10 μL reaction solution. The reaction mixture was run on a 4% native PAGE gel. e Quantification of the EMSA data shown in (d). The density of each RNA band was measured using Image Lab 6.0 (Bio-Rad), and the results were obtained from three independent experiments.