Fig. 5. Amino acids 461–463 in just one DGCR8 monomer are necessary for the accuracy and efficiency of microprocessor cleavage.
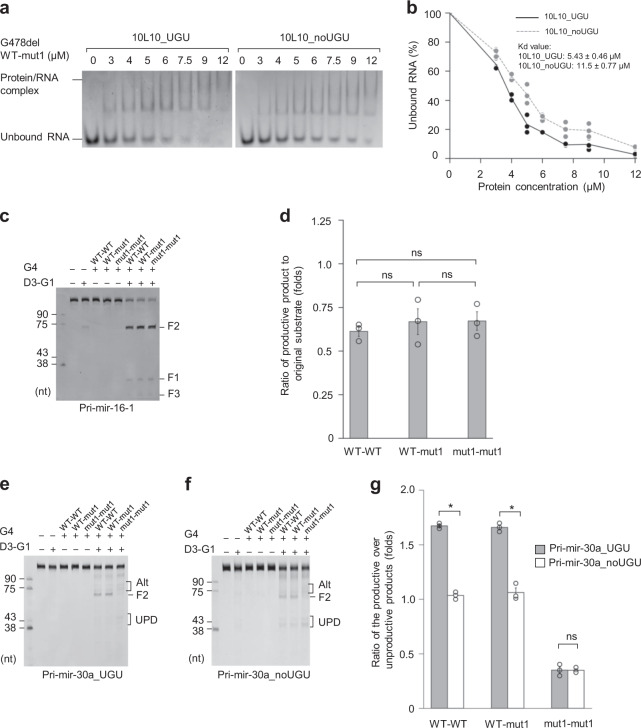
a EMSAs of the hybrid G478del WT–mut1 protein. Various amounts of G478del WT–mut1 (ranging from 0 to 12 μM) were mixed with 1 μM of either 10L10_UGU or 10L10_noUGU RNA in a 10 μL reaction solution. b Quantification of the EMSA data shown in (a). The density of each RNA band was determined using Image Lab 6.0 (Bio-Rad), and the results were obtained from three independent experiments. c The pri-mir-16-1 processing by microprocessor. Pri-mir-16-1 (0.5 μM) was processed by 0.1 μM D3–G1 and 0.05 μM G4 WT–WT, G4 WT–mut1, or G4 mut1–mut1 in the reaction buffer for 60 min, as described in “Methods”. d Quantification of the data shown in (c). The estimated ratio of the productive product (F2) to the original substrate was calculated from three independent experiments. G4 WT–WT vs. G4 WT–mut1: p = 0.534, G4 WT–mut1 vs. G4 mut1–mut1: p = 0.875, G4 WT–WT vs. G4 mut1–mut1: p = 0.373. e, f The pri-mir-30a_UGU (e) and pri-mir-30a_noUGU (f) processing by Microprocessor. Pri-mir-30a_UGU or pri-mir-30a_noUGU (both at 0.5 μM) was processed by 0.2 μM D3–G1 and 0.1 μM of G4 WT–WT, G4 WT–mut1, or G4 mut1–mut1 in the reaction buffer for 60 min, as described in “Methods”. Alt and UPD mean the alternative and unproductive cleavages, respectively. g Quantification of the data shown in (e, f). The ratio of the productive (F2) to unproductive products was calculated from three independent experiments. Pri-mir-30a_UGU vs. pri-mir-30a_noUGU for G4 WT–WT: p = 0.002, G4 WT–mut1: p = 0.002, and G4 mut1–mut1: p = 0.951. The asterisks (*) and (ns) indicate statistically significant and nonsignificant differences, respectively, from the two-sided t test.