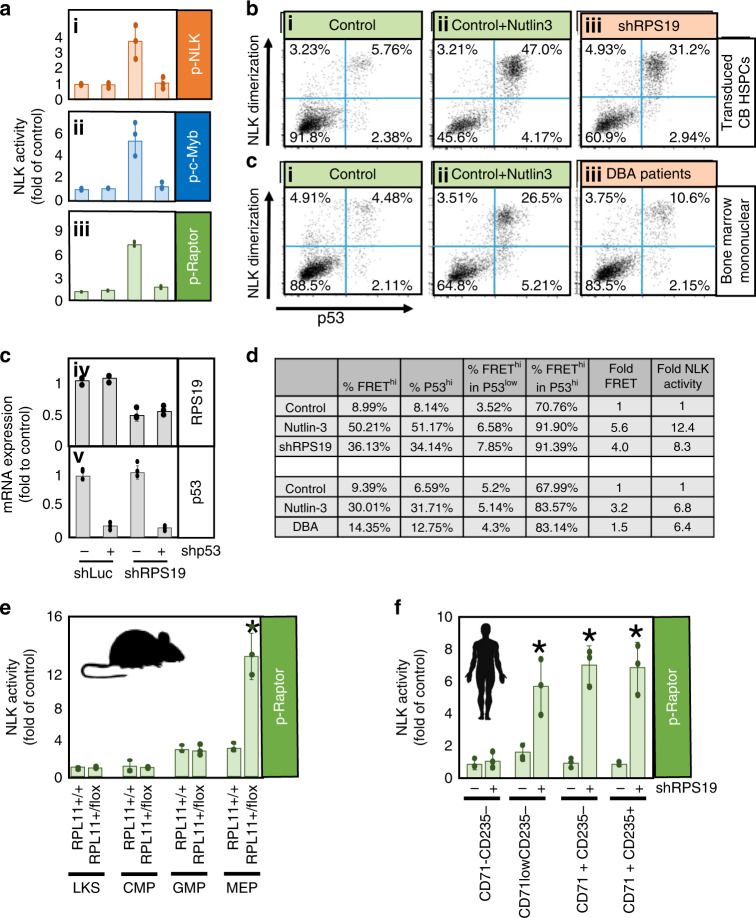
Fig. 6. NLK is activated in early erythroid progenitors and requires P53 stabilization in RPS19-insufficiency.
a Cord blood CD34+ progenitors were transduced with lentivirus expressing shRNA against luciferase (shLuc) or RPS19 (shRPS19) co-expressing GFP. Cells were also transduced with or without shRNA against p53 co-expressing mCherry. After 36 h, GFP+ and GFP+,mCherry+ cells were differentiated in erythroid media for 8 days, followed by NLK kinase assay (i—NLK: orange, ii—Myb: blue, iii—Raptor: green) and qRT-PCR analysis of RPS19 (iv) and p53 (v) expression. b CD34+ cord blood HSPCs were transduced with control shRNA (shLuc) along with YFP- and CFP-tagged NLK and allowed to differentiate for 3 days, in the presence (i) or absence (ii) of Nutlin-3. Cells were stained for p53 and both p53 and FRET was measured by flow cytometry. For comparison, HSPCs were transduced with shRNA against RPS19 (iii). c Bone marrow mononuclear cells from healthy donors were transduced with YFP- and CFP-NLK and incubated alone (i), or with Nutlin-3 (ii) for 24 h, prior to p53 and FRET analysis. Bone marrow mononuclear cells from three DBA patients were pooled and analyzed simultaneously (iii). d Documentation of the percentage of cells with dimerizing NLK in p53hi and p53low is tabulated and can be compared with NLK in vitro kinase activity. e—green Samples were gated to include non-erythroid CD71-CD235- populations, MEP- and BFU-E-enriched CD71lowCD235- populations, CFU-E-enriched CD71hiCD235- populations and proerythroblast and intermediate erythroblast CD71hiCD235+ populations. Sorted populations were lysed and immunoprecipitated NLK was subjected to in vitro kinase assay examining Raptor phosphorylation as a substrate. Activity was normalized to the non-erythroid control samples. f—green Sorted populations were lysed and immunoprecipitated NLK was subjected to in vitro kinase assay examining Raptor phosphorylation as a substrate. Activity was normalized to the Lin-Kit+Sca+ RPL11+/+ samples. Bars represent means ± SD with individual data points overlaid. n = 3 independent experiments performed in triplicate. Statistics: two-tailed Student’s t test, significant *p < 0.05.Also see Supplementary Fig. 7. Source data are provided as a Source Data file.