Figure 1.
LincRNA Expression Is Often Cell Cycle Regulated
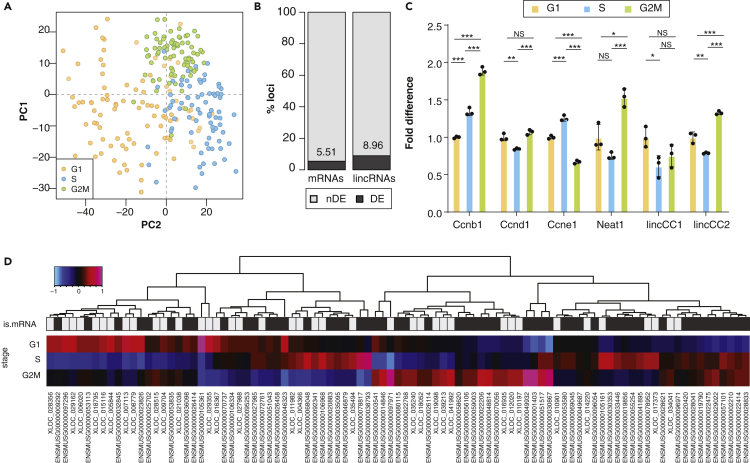
(A) Principal component analysis of gene expression for all loci passing the technical noise filter. The first two principal components (PC1 on the x axis and PC2 on the y axis) together explain 5.15% of the total variability, and separate cells in different cell cycle stages (G1: orange, S: blue; G2/M: green).
(B) Percentage of protein-coding genes (left) and lincRNAs (right) that are differentially expressed (dark gray, numbers indicate the percentage) between at least two cell cycle stages.
(C) Fold difference in normalized expression (relative to G1) of known cell cycle-regulated genes (Ccnb1, Ccnd1, Ccne1, Neat1) and two differentially expressed novel lincRNAs (lincCC1 and lincCC2) in mESCs at different cell cycle stages (G1: orange, S: blue, G2/M: green).
(D) Heatmap representing the median Z score of the shifted log expression level across all cells in each of the three stages (G1, S and G2M, rows) for each differentially expressed lincRNA and mRNA (columns). Rectangles in “is.mRNA” row are colored gray and black for lincRNA and mRNA, respectively. Color scale top right. Significance (two-tailed unpaired t test p value) between cell cycle stage expression is indicated as NS p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.