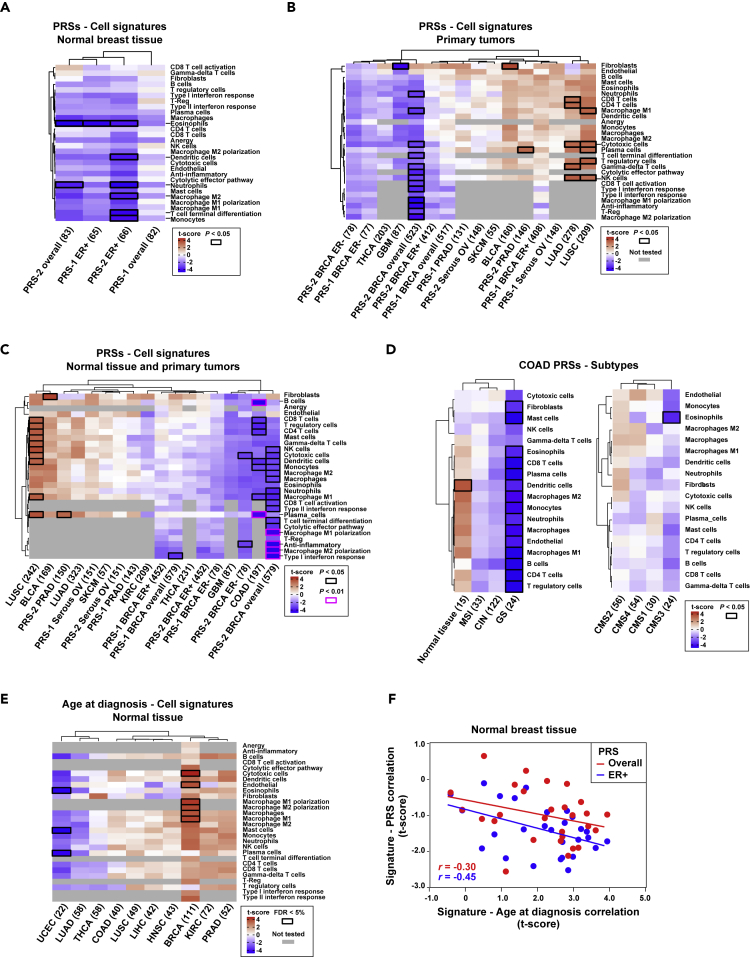
Figure 3.
Associations between Immune/Stromal Cell Signatures and PRSs
(A) Unsupervised clustering of the results of the regression analysis between cell signatures and PRSs in normal breast tissue. The y axis depicts the cell type signatures, and the x axis shows the PRSs. Sources #1 and #2 of the PRSs are detailed in Methods. ER+ and ER− indicate estrogen receptor-positive and estrogen receptor-negative subsets, respectively. The maximum sample size used in each analysis is shown. The color scale (t-score) is calculated as the β estimate divided by the standard error. Nominally significant associations are indicated by black-outlined rectangles.
(B) Unsupervised clustering of the coefficients of the regression of cell signature values in primary tumor TCGA studies and the corresponding PRSs. The gray-filled rectangles indicate “not tested” correlations because the corresponding cell signatures were only defined for breast cancer.
(C) Unsupervised clustering of the coefficients of the regression of cell signature values in combined normal tissue and primary tumor datasets, and the corresponding PRSs. The regression p values <0.01 are also indicated as depicted in the inset.
(D) Unsupervised clustering of the coefficients of the regression analysis between PRSs and cell signatures in combined normal tissue and primary tumors of the COAD study, divided by cancer subtypes.
(E) Unsupervised clustering of the results of the regression analysis of cell signatures in normal tissue and age at diagnosis across TCGA studies. Associations significant at a false discovery rate (FDR) < 5% are indicated by black-outlined rectangles.
(F) Negative correlations between the coefficients of regressions of immune/stromal cell contents and age at diagnosis or PRSs in normal breast tissue, for all cases and only ER-positive cases. The correlation coefficients are shown.