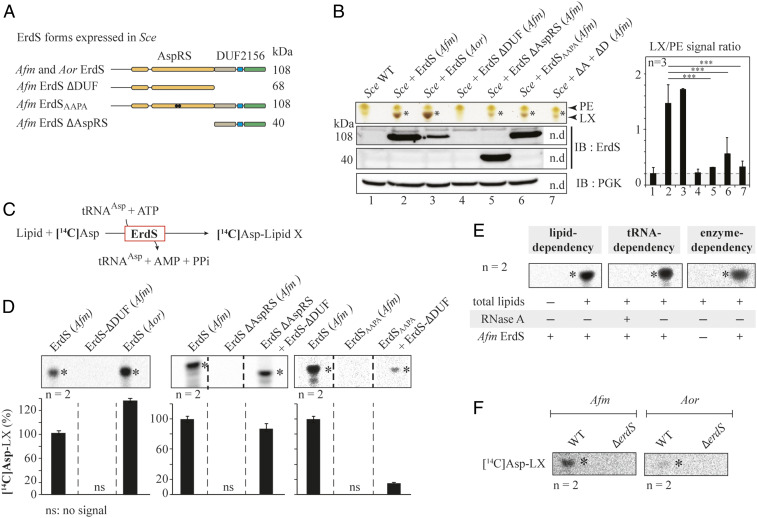
Fig. 3.
Dissecting ErdS lipid modification mechanism. (A) Schematized modular organization of full-length ErdS (108 kDa, Afm and Aor) and of its variants (Afm) that were expressed in the Sce heterologous model. ErdS-ΔDUF: AspRS standalone domain; ErdS-ΔAspRS: DUF2156 standalone domain (40 kDa); ErdSAAPA: catalytic null of the AspRS moiety (108 kDa). (B) TLC-based analysis of total lipids from Sce expressing ErdS variants described in A and Sce + (ΔD + ΔA) corresponds to the double expression of ErdS-ΔDUF + ErdS-ΔAspRS in Sce. The TLC plate stained with sulfuric acid/MnCl2 was cropped to the area of interest. LX signal (number of pixels) was normalized to that of PE in each case, and the LX/PE ratio is represented in a graph (Right). In the absence of LX, PG (phosphatidylglycerol) becomes visible; thus the PG/LX ratio obtained was considered “background signal” (gray background). However, the brown staining of LX (yellow for PG) made it possible to visually assess the presence of LX even for low LX/PE values. The Student's t test was used to assess the significance of the means of the data; ***P < 0.005. ErdS variants expression was analyzed by Western blot with an anti-Afm DUF2156 polyclonal antibodies (IB:ErdS), and loading control was performed with anti-PGK antibodies (IB:PGK). (C) Schematized reaction of the LA assay, described in Materials and Methods. (D) LX synthesis by the purified recombinant full-length and mutant ErdSs was measured using the LA assay. When mixed, proteins were in equimolar ratios. The [14C]-Asp lipid levels (percent) are provided below each TLC. (E) Verification of tRNA, lipid, and ErdS dependency of LX synthesis using LA assay; +: presence; −: absence. (F) Measurements of lipid X synthesis (ErdS activity) by WT and ΔerdS Aspergillus spp. crude extracts using LA assay; *: LX. The [14C]Asp lipids were revealed using phosphorimaging (D–F). TLCs and immunoblots are representative of two to three independent experiments, and the number of replicates (n) is indicated.